Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner
- PMID: 17981978
- PMCID: PMC2223732
- DOI: 10.1128/JB.00826-07
Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner
Abstract
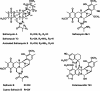
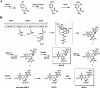
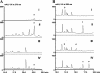
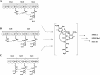
Saframycin A (SFM-A), produced by Streptomyces lavendulae NRRL 11002, belongs to the tetrahydroisoquinoline family of antibiotics, and its core is structurally similar to the core of ecteinascidin 743, which is a highly potent antitumor drug isolated from a marine tunicate. In this study, the biosynthetic gene cluster for SFM-A was cloned and localized to a 62-kb contiguous DNA region. Sequence analysis revealed 30 genes that constitute the SFM-A gene cluster, encoding an unusual nonribosomal peptide synthetase (NRPS) system and tailoring enzymes and regulatory and resistance proteins. The results of substrate prediction and in vitro characterization of the adenylation specificities of this NRPS system support the hypothesis that the last module acts in an iterative manner to form a tetrapeptidyl intermediate and that the colinearity rule does not apply. Although this mechanism is different from those proposed for the SFM-A analogs SFM-Mx1 and safracin B (SAC-B), based on the high similarity of these systems, it is likely they share a common mechanism of biosynthesis as we describe here. Construction of the biosynthetic pathway of SFM-Y3, an aminated SFM-A, was achieved in the SAC-B producer (Pseudomonas fluorescens). These findings not only shed new insight on tetrahydroisoquinoline biosynthesis but also demonstrate the feasibility of engineering microorganisms to generate structurally more complex and biologically more active analogs by combinatorial biosynthesis.
Figures


Similar articles
-
The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase.Chem Biol. 2000 Aug;7(8):623-42. doi: 10.1016/s1074-5521(00)00011-9. Chem Biol. 2000. PMID: 11048953
-
Biosynthesis of 3-hydroxy-5-methyl-o-methyltyrosine in the saframycin/ safracin biosynthetic pathway.J Microbiol Biotechnol. 2009 May;19(5):439-46. doi: 10.4014/jmb.0808.484. J Microbiol Biotechnol. 2009. PMID: 19494690
-
Identification and characterization of the pyridomycin biosynthetic gene cluster of Streptomyces pyridomyceticus NRRL B-2517.J Biol Chem. 2011 Jun 10;286(23):20648-57. doi: 10.1074/jbc.M110.180000. Epub 2011 Mar 22. J Biol Chem. 2011. PMID: 21454714 Free PMC article.
-
Biosynthesis of Tetrahydroisoquinoline Antibiotics.Curr Top Med Chem. 2016;16(15):1717-26. doi: 10.2174/1568026616666151012112329. Curr Top Med Chem. 2016. PMID: 26456466 Review.
-
The biosynthetic gene cluster for the anticancer drug bleomycin from Streptomyces verticillus ATCC15003 as a model for hybrid peptide-polyketide natural product biosynthesis.J Ind Microbiol Biotechnol. 2001 Dec;27(6):378-85. doi: 10.1038/sj.jim.7000194. J Ind Microbiol Biotechnol. 2001. PMID: 11774003 Review.
Cited by
-
Exploring the Diversity and Specificity of Secondary Biosynthetic Potential in Rhodococcus.Mar Drugs. 2024 Sep 6;22(9):409. doi: 10.3390/md22090409. Mar Drugs. 2024. PMID: 39330290 Free PMC article.
-
Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia.Appl Environ Microbiol. 2012 Jun;78(12):4468-80. doi: 10.1128/AEM.00823-12. Epub 2012 Apr 6. Appl Environ Microbiol. 2012. PMID: 22492438 Free PMC article.
-
Ketoreductase Domain Dysfunction Expands Chemodiversity: Malyngamide Biosynthesis in the Cyanobacterium Okeania hirsuta.ACS Chem Biol. 2018 Dec 21;13(12):3385-3395. doi: 10.1021/acschembio.8b00910. Epub 2018 Dec 3. ACS Chem Biol. 2018. PMID: 30444349 Free PMC article.
-
The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry.J Bacteriol. 2010 May;192(10):2583-95. doi: 10.1128/JB.01526-09. Epub 2010 Mar 19. J Bacteriol. 2010. PMID: 20304989 Free PMC article.
-
Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.Nat Commun. 2021 Dec 6;12(1):7085. doi: 10.1038/s41467-021-27404-3. Nat Commun. 2021. PMID: 34873166 Free PMC article.
References
-
- Ahlert, J., E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet, and J. S. Thorson. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 2971173-1176. - PubMed
-
- Arai, T., K. Takahashi, K. Ishiguro, and K. Yazawa. 1980. Increased production of saframycin A and isolation of saframycin S. J. Antibiot. 33951-960. - PubMed
-
- Arai, T., K. Takahashi, K. Ishiguro, and Y. Mikami. 1980. Some chemotherapeutic properties of two new antitumor antibiotics, saframycins A and C. Gann 71790-796. - PubMed
-
- Baltz, R. H. 2006. Molecular engineering approaches to peptide, polyketide and other antibiotics. Nat. Biotechnol. 241533-1540. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

