Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling
- PMID: 17981998
- PMCID: PMC2174870
- DOI: 10.1105/tpc.107.053371
Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling
Abstract
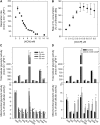
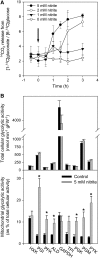
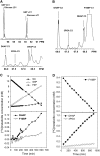
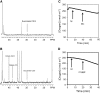
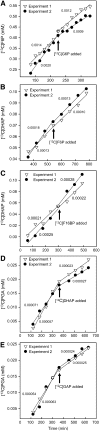
In Arabidopsis thaliana, enzymes of glycolysis are present on the surface of mitochondria and free in the cytosol. The functional significance of this dual localization has now been established by demonstrating that the extent of mitochondrial association is dependent on respiration rate in both Arabidopsis cells and potato (Solanum tuberosum) tubers. Thus, inhibition of respiration with KCN led to a proportional decrease in the degree of association, whereas stimulation of respiration by uncoupling, tissue ageing, or overexpression of invertase led to increased mitochondrial association. In all treatments, the total activity of the glycolytic enzymes in the cell was unaltered, indicating that the existing pools of each enzyme repartitioned between the cytosol and the mitochondria. Isotope dilution experiments on isolated mitochondria, using (13)C nuclear magnetic resonance spectroscopy to monitor the impact of unlabeled glycolytic intermediates on the production of downstream intermediates derived from (13)C-labeled precursors, provided direct evidence for the occurrence of variable levels of substrate channeling. Pull-down experiments suggest that interaction with the outer mitochondrial membrane protein, VDAC, anchors glycolytic enzymes to the mitochondrial surface. It appears that glycolytic enzymes associate dynamically with mitochondria to support respiration and that substrate channeling restricts the use of intermediates by competing metabolic pathways.
Figures

References
-
- Averill, R.H., Bailey-Serres, J., and Kruger, N.J. (1998). Co-operation between cytosolic and plastidic oxidative pentose phosphate pathways revealed by 6-phosphogluconate dehydrogenase-deficient genotypes of maize. Plant J. 14 449–457.
-
- Cho, Y.H., Yoo, S.D., and Sheen, J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589. - PubMed
-
- Claeyssen, E., and Rivoal, J. (2007). Isozymes of plant hexokinase: Occurrence, properties and functions. Phytochemistry 68 709–731. - PubMed
-
- Clegg, J.S., and Jackson, S.A. (1990). Glucose metabolism and the channeling of glycolytic intermediates in permeabilized L-929 cells. Arch. Biochem. Biophys. 278 452–460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

