Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH
- PMID: 17982447
- PMCID: PMC2679367
- DOI: 10.1038/nchembio.2007.49
Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH
Abstract
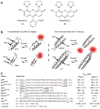
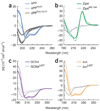
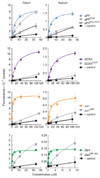
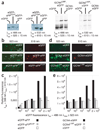
Recombinant polypeptides and protein domains containing two cysteine pairs located distal in primary sequence but proximal in the native folded or assembled state are labeled selectively in vitro and in mammalian cells using the profluorescent biarsenical reagents FlAsH-EDT2 and ReAsH-EDT2. This strategy, termed bipartite tetracysteine display, enables the detection of protein-protein interactions and alternative protein conformations in live cells. As proof of principle, we show that the equilibrium stability and fluorescence intensity of polypeptide-biarsenical complexes correlates with the thermodynamic stability of the protein fold or assembly. Destabilized protein variants form less stable and less bright biarsenical complexes, which allows discrimination of live cells expressing folded polypeptide and protein domains from those containing disruptive point mutations. Bipartite tetracysteine display may provide a means to detect early protein misfolding events associated with Alzheimer's disease, Parkinson's disease and cystic fibrosis; it may also enable high-throughput screening of compounds that stabilize discrete protein folds.
Figures

References
-
- Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. - PubMed
-
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green fluorescent protein. Gene. 1992;111:229–233. - PubMed
-
- Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. - PubMed
-
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. - PubMed
-
- Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- PubChem-Substance/26738251
- PubChem-Substance/26738252
- PubChem-Substance/26738253
- PubChem-Substance/26738254
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

