Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme
- PMID: 17984226
- PMCID: PMC2223317
- DOI: 10.1128/MCB.01168-07
Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme
Abstract
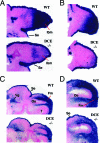
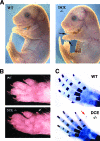
Tcfap2a, the gene encoding the mouse AP-2alpha transcription factor, is required for normal development of multiple structures during embryogenesis, including the face and limbs. Using comparative sequence analysis and transgenic-mouse experiments we have identified an intronic enhancer within this gene that directs expression to the face and limb mesenchyme. There are two conserved sequence blocks within this intron, and the larger of these directs tissue-specific activity and is found in all vertebrate Tcfap2a genes analyzed. To assess the role of the enhancer in regulating endogenous mouse Tcfap2a expression, we have deleted this cis-regulatory sequence from the genome. Loss of this element severely impairs Tcfap2a expression in the limb bud mesenchyme but generates only a modest reduction in the facial mesenchyme. The reduction in Tcfap2a transcription is accompanied by altered patterning of the forelimb, resulting in postaxial polydactyly. These results indicate that the major role for this enhancer resides within the limb bud, and it serves to maintain a level of Tcfap2a expression that limits the size of the hand plate and the associated number of digit primordia. The potential role of this cis-acting sequence in modeling the size and shape of the face and limbs during evolution is discussed.
Figures


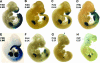


References
-
- Ahituv, N., A. Erven, H. Fuchs, K. Guy, R. Ashery-Padan, T. Williams, M. H. de Angelis, K. B. Avraham, and K. P. Steel. 2004. An ENU-induced mutation in AP-2α leads to middle ear and ocular defects in Doarad mice. Mamm. Genome 15424-432. - PubMed
-
- Boffelli, D., M. A. Nobrega, and E. M. Rubin. 2004. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 5456-465. - PubMed
-
- Brewer, S., W. Feng, J. Huang, S. Sullivan, and T. Williams. 2004. Wnt1-Cre mediated deletion of AP-2α causes multiple neural crest related defects. Dev. Biol. 267135-152. - PubMed
-
- Brewer, S., X. Jiang, S. Donaldson, T. Williams, and H. M. Sucov. 2002. Requirement for AP-2α in cardiac outflow tract morphogenesis. Mech. Dev. 110139-149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
