Exonuclease removal of dideoxycytidine (zalcitabine) by the human mitochondrial DNA polymerase
- PMID: 17984232
- PMCID: PMC2223897
- DOI: 10.1128/AAC.00778-07
Exonuclease removal of dideoxycytidine (zalcitabine) by the human mitochondrial DNA polymerase
Abstract
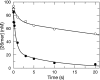
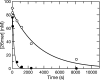
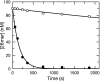
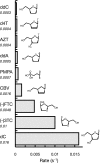
The toxicity of nucleoside analogs used for the treatment of human immunodeficiency virus infection is due primarily to the inhibition of replication of the mitochondrial genome by the human mitochondrial DNA polymerase (Pol gamma). The severity of clinically observed toxicity correlates with the kinetics of incorporation versus excision of each analog as quantified by a toxicity index, spanning over six orders of magnitude. Here we show that the rate of excision of dideoxycytidine (zalcitabine; ddC) was reduced fourfold (giving a half-life of approximately 2.4 h) by the addition of a physiological concentration of deoxynucleoside triphosphates (dNTPs) due to the formation of a tight ternary enzyme-DNA-dNTP complex at the polymerase site. In addition, we provide a more accurate measurement of the rate of excision and show that the low rate of removal of ddCMP results from both the unfavorable transfer of the primer strand from the polymerase to the exonuclease site and the inefficient binding and/or hydrolysis at the exonuclease site. The analogs ddC, stavudine, and ddATP (a metabolite of didanosine) each bind more tightly at the polymerase site during incorporation than normal nucleotides, and this tight binding contributes to slower excision by the proofreading exonuclease, leading to increased toxicity toward mitochondrial DNA.
Figures
References
-
- Adkins, J. C., D. H. Peters, and D. Faulds. 1997. Zalcitabine. An update of its pharmacodynamic and pharmacokinetic properties and clinical efficacy in the management of HIV infection. Drugs 53:1054-1080. - PubMed
-
- Cammack, N., P. Rouse, C. L. Marr, P. J. Reid, R. E. Boehme, J. A. Coates, C. R. Penn, and J. M. Cameron. 1992. Cellular metabolism of (−) enantiomeric 2′-deoxy-3′-thiacytidine. Biochem. Pharmacol. 43:2059-2064. - PubMed
-
- Chang, C. N., S. L. Doong, J. H. Zhou, J. W. Beach, L. S. Jeong, C. K. Chu, C. H. Tsai, Y. C. Cheng, D. Liotta, and R. Schinazi. 1992. Deoxycytidine deaminase-resistant stereoisomer is the active form of (+/−)-2′,3′-dideoxy-3′-thiacytidine in the inhibition of hepatitis B virus replication. J. Biol. Chem. 267:13938-13942. - PubMed
-
- Chang, C. N., V. Skalski, J. H. Zhou, and Y. C. Cheng. 1992. Biochemical pharmacology of (+)- and (−)-2′,3′-dideoxy-3′-thiacytidine as anti-hepatitis B virus agents. J. Biol. Chem. 267:22414-22420. - PubMed
-
- Donlin, M. J., S. S. Patel, and K. A. Johnson. 1991. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry 30:538-546. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

