Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin
- PMID: 17984306
- PMCID: PMC2118507
- DOI: 10.1084/jem.20071082
Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin
Abstract
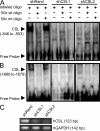
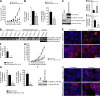
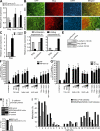
Aberrant expression of Jagged1 and Notch1 are associated with poor outcome in breast cancer. However, the reason that Jagged1 and/or Notch overexpression portends a poor prognosis is unknown. We identify Slug, a transcriptional repressor, as a novel Notch target and show that elevated levels of Slug correlate with increased expression of Jagged1 in various human cancers. Slug was essential for Notch-mediated repression of E-cadherin, which resulted in beta-catenin activation and resistance to anoikis. Inhibition of ligand-induced Notch signaling in xenografted Slug-positive/E-cadherin-negative breast tumors promoted apoptosis and inhibited tumor growth and metastasis. This response was associated with down-regulated Slug expression, reexpression of E-cadherin, and suppression of active beta-catenin. Our findings suggest that ligand-induced Notch activation, through the induction of Slug, promotes tumor growth and metastasis characterized by epithelial-to-mesenchymal transition and inhibition of anoikis.
Figures


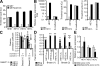
References
-
- Mumm, J.S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151–165. - PubMed
-
- Leong, K.G., and A. Karsan. 2006. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 107:2223–2233. - PubMed
-
- Gallahan, D., C. Jhappan, G. Robinson, L. Hennighausen, R. Sharp, E. Kordon, R. Callahan, G. Merlino, and G.H. Smith. 1996. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 56:1775–1785. - PubMed
-
- Callahan, R., and S.E. Egan. 2004. Notch signaling in mammary development and oncogenesis. J. Mammary Gland Biol. Neoplasia. 9:145–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

