Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring
- PMID: 17986188
- PMCID: PMC4764085
- DOI: 10.1111/j.1365-2958.2007.05998.x
Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring
Abstract
In bacteria, the actin-like FtsA protein interacts with the tubulin-like FtsZ protein, helping to assemble the cytokinetic Z ring, anchor it to the cytoplasmic membrane and recruit other essential divisome proteins. FtsA also interacts with itself, but it is not clear whether this self-interaction is required for its full functionality. Here we describe new dominant negative missense mutations in Escherichia coli ftsA that specifically inhibit FtsA homodimerization and simultaneously cause disruption of Z rings. The negative effects of one mutation, M71A, were suppressed by altering levels of certain division proteins or by additional mutations in ftsA that promote increased integrity of the Z ring. Remarkably, when FtsA, FtsA-M71A, and other mutants of FtsA that compromise self-interaction were connected in a tandem repeat, they were at least partially functional and suppressed defects of an ftsZ84(ts) mutation. This gain of function by FtsA tandems further suggested that FtsA monomers cause deleterious interactions with FtsZ and that increased dimerization or oligomerization of FtsA enhances its ability to promote Z-ring integrity. Therefore, we propose that FtsZ assembly is regulated by the extent of FtsA oligomerization.
Figures
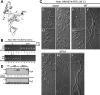

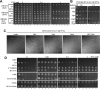
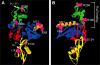

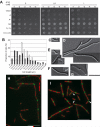

Similar articles
-
FtsA Regulates Z-Ring Morphology and Cell Wall Metabolism in an FtsZ C-Terminal Linker-Dependent Manner in Caulobacter crescentus.J Bacteriol. 2020 Mar 11;202(7):e00693-19. doi: 10.1128/JB.00693-19. Print 2020 Mar 11. J Bacteriol. 2020. PMID: 31932314 Free PMC article.
-
Assembly and architecture of Escherichia coli divisome proteins FtsA and FtsZ.J Biol Chem. 2022 Mar;298(3):101663. doi: 10.1016/j.jbc.2022.101663. Epub 2022 Jan 29. J Biol Chem. 2022. PMID: 35104502 Free PMC article.
-
FtsEX acts on FtsA to regulate divisome assembly and activity.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):E5052-61. doi: 10.1073/pnas.1606656113. Epub 2016 Aug 8. Proc Natl Acad Sci U S A. 2016. PMID: 27503875 Free PMC article.
-
Building the Bacterial Divisome at the Septum.Subcell Biochem. 2024;104:49-71. doi: 10.1007/978-3-031-58843-3_4. Subcell Biochem. 2024. PMID: 38963483 Review.
-
FtsZ-ring Architecture and Its Control by MinCD.Subcell Biochem. 2017;84:213-244. doi: 10.1007/978-3-319-53047-5_7. Subcell Biochem. 2017. PMID: 28500527 Review.
Cited by
-
A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling.Mol Microbiol. 2015 Sep;97(5):988-1005. doi: 10.1111/mmi.13081. Epub 2015 Jul 4. Mol Microbiol. 2015. PMID: 26046682 Free PMC article.
-
Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling.Mol Microbiol. 2018 Sep;109(5):676-693. doi: 10.1111/mmi.14069. Epub 2018 Aug 1. Mol Microbiol. 2018. PMID: 29995995 Free PMC article.
-
FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins.Mol Microbiol. 2012 Jan;83(1):151-67. doi: 10.1111/j.1365-2958.2011.07923.x. Epub 2011 Nov 29. Mol Microbiol. 2012. PMID: 22111832 Free PMC article.
-
Alp7R regulates expression of the actin-like protein Alp7A in Bacillus subtilis.J Bacteriol. 2012 May;194(10):2715-24. doi: 10.1128/JB.06550-11. Epub 2012 Mar 16. J Bacteriol. 2012. PMID: 22427628 Free PMC article.
-
The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN.J Bacteriol. 2012 Apr;194(8):1989-2000. doi: 10.1128/JB.06683-11. Epub 2012 Feb 10. J Bacteriol. 2012. PMID: 22328664 Free PMC article.
References
-
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. - PubMed
-
- Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 1997;179:4277–4284. - PMC - PubMed
-
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

