Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures
- PMID: 17986617
- PMCID: PMC2084285
- DOI: 10.1073/pnas.0705038104
Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures
Abstract
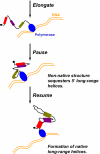
RNA folding in the cell occurs during transcription. Expedient RNA folding must avoid the formation of undesirable structures as the nascent RNA emerges from the RNA polymerase. We show that efficient folding during transcription of three conserved noncoding RNAs from Escherichia coli, RNase P RNA, signal-recognition particle RNA, and tmRNA is facilitated by their cognate polymerase pausing at specific locations. These pause sites are located between the upstream and downstream portions of all of the native long-range helices in these noncoding RNAs. In the paused complexes, the nascent RNAs form labile structures that sequester these upstream portions in a manner to possibly guide folding. Both the pause sites and the secondary structure of the nonnative portions of the paused complexes are phylogenetically conserved among gamma-proteobacteria. We propose that specific pausing-induced structural formation is a general strategy to facilitate the folding of long-range helices. This polymerase-based mechanism may result in portions of noncoding RNA sequences being evolutionarily conserved for efficient folding during transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

