Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells
- PMID: 17988662
- PMCID: PMC2174610
- DOI: 10.1016/j.ydbio.2007.09.057
Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells
Abstract
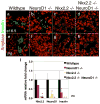
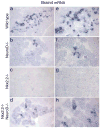
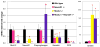
Nkx2.2 and NeuroD1 are vital for proper differentiation of pancreatic islet cell types. Nkx2.2-null mice fail to form beta cells, have reduced numbers of alpha and PP cells and display an increase in ghrelin-producing epsilon cells. NeuroD1-null mice display a reduction of alpha and beta cells after embryonic day (e) 17.5. To begin to determine the relative contributions of Nkx2.2 and NeuroD1 in islet development, we generated Nkx2.2-/-;NeuroD1-/- double knockout (DKO) mice. As expected, the DKO mice fail to form beta cells, similar to the Nkx2.2-null mice, suggesting that the Nkx2.2 phenotype may be dominant over the NeuroD1 phenotype in the beta cells. Surprisingly, however, the alpha, PP and epsilon phenotypes of the Nkx2.2-null mice are partially rescued by the simultaneous elimination of NeuroD1, even at early developmental time points when NeuroD1 null mice alone do not display a phenotype. Our results indicate that Nkx2.2 and NeuroD1 interact to regulate pancreatic islet cell fates, and this epistatic relationship is cell-type dependent. Furthermore, this study reveals a previously unappreciated early function of NeuroD1 in regulating the specification of alpha, PP and epsilon cells.
Figures




References
-
- Cerf ME, et al. Transcription factors, pancreatic development, and beta-cell maintenance. Biochem Biophys Res Commun. 2005;326:699–702. - PubMed
-
- Chu K, et al. BETA2 and pancreatic islet development. Recent Prog Horm Res. 2001;56:23–46. - PubMed
-
- Collombat P, et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

