NHA-oc/NHA2: a mitochondrial cation-proton antiporter selectively expressed in osteoclasts
- PMID: 17988971
- PMCID: PMC3593247
- DOI: 10.1016/j.bone.2007.09.046
NHA-oc/NHA2: a mitochondrial cation-proton antiporter selectively expressed in osteoclasts
Abstract
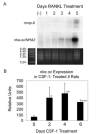
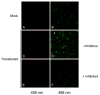
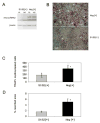
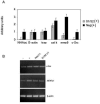
Bone resorption is regulated by a complex system of hormones and cytokines that cause osteoblasts/stromal cells and lymphocytes to produce factors including RANKL, that ultimately result in the differentiation and activation of osteoclasts, the bone resorbing cells. We used a microarray approach to identify genes upregulated in RANKL-stimulated osteoclast precursor cells. Osteoclast expression was confirmed by multiple tissue Northern and in situ hybridization analysis. Gene function studies were carried out by siRNA analysis. We identified a novel gene, which we termed nha-oc/NHA2, which is strongly upregulated during RANKL-induced osteoclast differentiation in vitro and in vivo. nha-oc/NHA2 encodes a novel cation-proton antiporter (CPA) and is the mouse orthologue of a human gene identified in a database search: HsNHA2. nha-oc/NHA2 is selectively expressed in osteoclasts. NHA-oc/NHA2 protein localizes to the mitochondria, where it mediates Na(+)-dependent changes in mitochondrial pH and Na(+) acetate induced mitochondrial passive swelling. RNA silencing of nha-oc/nha2 reduces osteoclast differentiation and resorption, suggesting a role for NHA-oc/NHA2 in these processes. nha-oc/NHA2 therefore is a novel member of the CPA family and is the first mitochondrial NHA characterized to date. nha-oc/NHA2 is also unique in that it is the first eukaryotic and tissue-specific CPA2 characterized to date. NHA-oc/NHA2 displays the expected activities of a bona fide CPA and plays a key role(s) in normal osteoclast differentiation and function.
Figures






Similar articles
-
Expression analysis of nha-oc/NHA2: a novel gene selectively expressed in osteoclasts.Gene Expr Patterns. 2007 Oct;7(8):846-51. doi: 10.1016/j.modgep.2007.07.002. Epub 2007 Aug 14. Gene Expr Patterns. 2007. PMID: 17698421 Free PMC article.
-
Sodium/hydrogen exchanger NHA2 in osteoclasts: subcellular localization and role in vitro and in vivo.Bone. 2010 Aug;47(2):331-40. doi: 10.1016/j.bone.2010.04.605. Epub 2010 May 11. Bone. 2010. PMID: 20441802
-
Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation.J Cell Physiol. 2008 May;215(2):497-505. doi: 10.1002/jcp.21331. J Cell Physiol. 2008. PMID: 18064667 Free PMC article.
-
Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families.Endocr Rev. 1999 Jun;20(3):345-57. doi: 10.1210/edrv.20.3.0367. Endocr Rev. 1999. PMID: 10368775 Review.
-
Osteoclast differentiation by RANKL and OPG signaling pathways.J Bone Miner Metab. 2021 Jan;39(1):19-26. doi: 10.1007/s00774-020-01162-6. Epub 2020 Oct 20. J Bone Miner Metab. 2021. PMID: 33079279 Review.
Cited by
-
Secretory Carrier Membrane Protein 2 Regulates Cell-surface Targeting of Brain-enriched Na+/H+ Exchanger NHE5.J Biol Chem. 2009 May 15;284(20):13892-13903. doi: 10.1074/jbc.M807055200. Epub 2009 Mar 10. J Biol Chem. 2009. PMID: 19276089 Free PMC article.
-
Melphalan flufenamide inhibits osteoclastogenesis by suppressing proliferation of monocytes.Bone Rep. 2021 Jun 7;15:101098. doi: 10.1016/j.bonr.2021.101098. eCollection 2021 Dec. Bone Rep. 2021. PMID: 34150958 Free PMC article.
-
Ethyl isopropyl amiloride decreases oxidative phosphorylation and increases mitochondrial fusion in clonal untransformed and cancer cells.Am J Physiol Cell Physiol. 2021 Jul 1;321(1):C147-C157. doi: 10.1152/ajpcell.00001.2021. Epub 2021 May 26. Am J Physiol Cell Physiol. 2021. PMID: 34038242 Free PMC article.
-
Loss of Sodium/Hydrogen Exchanger NHA2 Exacerbates Obesity- and Aging-Induced Glucose Intolerance in Mice.PLoS One. 2016 Sep 29;11(9):e0163568. doi: 10.1371/journal.pone.0163568. eCollection 2016. PLoS One. 2016. PMID: 27685945 Free PMC article.
-
PAMM: a redox regulatory protein that modulates osteoclast differentiation.Antioxid Redox Signal. 2010 Jul 1;13(1):27-37. doi: 10.1089/ars.2009.2886. Antioxid Redox Signal. 2010. PMID: 19951071 Free PMC article.
References
-
- Burckhardt G, Di Sole F, Helmle-Kolb C. The Na+/H+ exchanger gene family. J Nephrol. 2002;15(5):S3–21. - PubMed
-
- Demaurex N, Grinstein S. Na+/H+ antiport: modulation by ATP and role in cell volume regulation. J Exp Biol. 1994;196:389–404. - PubMed
-
- Ritter M, Fuerst J, Woll E, et al. Na(+)/H(+)exchangers: linking osmotic dysequilibrium to modified cell function. Cell Physiol Biochem. 2001;11:1–18. - PubMed
-
- Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–C239. - PubMed
-
- Chang AB, Lin R, Keith SW, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

