Significant gene content variation characterizes the genomes of inbred mouse strains
- PMID: 17989247
- PMCID: PMC2099583
- DOI: 10.1101/gr.6754607
Significant gene content variation characterizes the genomes of inbred mouse strains
Abstract
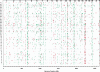
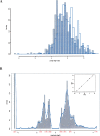
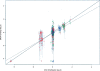
The contribution to genetic diversity of genomic segmental copy number variations (CNVs) is less well understood than that of single-nucleotide polymorphisms (SNPs). While less frequent than SNPs, CNVs have greater potential to affect phenotype. In this study, we have performed the most comprehensive survey to date of CNVs in mice, analyzing the genomes of 42 Mouse Phenome Consortium priority strains. This microarray comparative genomic hybridization (CGH)-based analysis has identified 2094 putative CNVs, with an average of 10 Mb of DNA in 51 CNVs when individual mouse strains were compared to the reference strain C57BL/6J. This amount of variation results in gene content that can differ by hundreds of genes between strains. These genes include members of large families such as the major histocompatibility and pheromone receptor genes, but there are also many singleton genes including genes with expected phenotypic consequences from their deletion or amplification. Using a whole-genome association analysis, we demonstrate that complex multigenic phenotypes, such as food intake, can be associated with specific copy number changes.
Figures



Similar articles
-
A high-resolution map of segmental DNA copy number variation in the mouse genome.PLoS Genet. 2007 Jan 5;3(1):e3. doi: 10.1371/journal.pgen.0030003. Epub 2006 Nov 22. PLoS Genet. 2007. PMID: 17206864 Free PMC article.
-
Copy number variation influences gene expression and metabolic traits in mice.Hum Mol Genet. 2009 Nov 1;18(21):4118-29. doi: 10.1093/hmg/ddp360. Epub 2009 Jul 31. Hum Mol Genet. 2009. PMID: 19648292 Free PMC article.
-
Genomic landscapes of endogenous retroviruses unveil intricate genetics of conventional and genetically-engineered laboratory mouse strains.Exp Mol Pathol. 2016 Apr;100(2):248-56. doi: 10.1016/j.yexmp.2016.01.005. Epub 2016 Jan 11. Exp Mol Pathol. 2016. PMID: 26779669 Free PMC article.
-
Copy number variations and clinical cytogenetic diagnosis of constitutional disorders.Nat Genet. 2007 Jul;39(7 Suppl):S48-54. doi: 10.1038/ng2092. Nat Genet. 2007. PMID: 17597782 Review.
-
Methods and strategies for analyzing copy number variation using DNA microarrays.Nat Genet. 2007 Jul;39(7 Suppl):S16-21. doi: 10.1038/ng2028. Nat Genet. 2007. PMID: 17597776 Free PMC article. Review.
Cited by
-
Genome-wide identification of copy number variations in Chinese Holstein.PLoS One. 2012;7(11):e48732. doi: 10.1371/journal.pone.0048732. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144949 Free PMC article.
-
Analysis of copy number variations among diverse cattle breeds.Genome Res. 2010 May;20(5):693-703. doi: 10.1101/gr.105403.110. Epub 2010 Mar 8. Genome Res. 2010. PMID: 20212021 Free PMC article.
-
Copy number variation in the bovine genome.BMC Genomics. 2010 May 6;11:284. doi: 10.1186/1471-2164-11-284. BMC Genomics. 2010. PMID: 20459598 Free PMC article.
-
Coding of pheromones by vomeronasal receptors.Cell Tissue Res. 2021 Jan;383(1):367-386. doi: 10.1007/s00441-020-03376-6. Epub 2021 Jan 12. Cell Tissue Res. 2021. PMID: 33433690 Review.
-
Next-generation sequencing of experimental mouse strains.Mamm Genome. 2012 Oct;23(9-10):490-8. doi: 10.1007/s00335-012-9402-6. Epub 2012 Jul 7. Mamm Genome. 2012. PMID: 22772437 Free PMC article. Review.
References
-
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. - PMC - PubMed
-
- Barrett M.T., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Tsang P., Curry B., Baird K., Meltzer P.S., Curry B., Baird K., Meltzer P.S., Baird K., Meltzer P.S., Meltzer P.S., et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl. Acad. Sci. 2004;101:17765–17770. - PMC - PubMed
-
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Eppig J.T., Festing M.F., Fisher E.M., Festing M.F., Fisher E.M., Fisher E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. - PubMed
-
- Behringer R.R., Nagy A., Gerstenstein M., Vintersten K., Nagy A., Gerstenstein M., Vintersten K., Gerstenstein M., Vintersten K., Vintersten K. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003.
-
- Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Coates G., Cuff J., Curwen V., Cutts T., Cuff J., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. An overview of Ensembl. Genome Res. 2004;14:925–928. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
