RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans
- PMID: 17989692
- PMCID: PMC2140109
- DOI: 10.1038/sj.emboj.7601922
RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans
Abstract
The small Ras-like GTPase Rap1 has been identified as a regulator of integrin activation and cadherin-mediated cell-cell contacts. Surprisingly, null mutants of RAP-1 in Caenorhabditis elegans are viable and fertile. In a synthetic lethal RNAi screen with C. elegans rap-1 mutants, the Ras-like GTPase ral-1 emerged as one of seven genes specifically required for viability. Depletion of exoc-8 and sec-5, encoding two putative RAL-1 effectors and members of the exocyst complex, also caused lethality of rap-1 mutants, but did not affect wild-type worms. The RAP-1 and the RAL-1/exocyst pathway appear to coordinate hypodermal cell movement and elongation during embryonic development. They mediate their effect in part through targeting the alpha-catenin homologue HMP-1 to the lateral membrane. Genetic interactions show that the RAP-1 and RAL-1/exocyst pathway also act in parallel during larval stages. Together these data provide in vivo evidence for the exocyst complex as a downstream RAL-1 effector in cell migration.
Figures


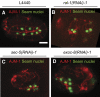
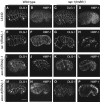

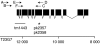
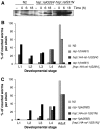
References
-
- Bos JL (2005) Linking Rap to cell adhesion. Curr Opin Cell Biol 17: 123–128 - PubMed
-
- Bossinger O, Klebes A, Segbert C, Theres C, Knust E (2001) Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev Biol 230: 29–42 - PubMed
-
- Bryant DM, Stow JL (2004) The ins and outs of E-cadherin trafficking. Trends Cell Biol 14: 427–434 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

