Moving forward in colorectal cancer research, what proteomics has to tell
- PMID: 17990347
- PMCID: PMC4205428
- DOI: 10.3748/wjg.v13.i44.5813
Moving forward in colorectal cancer research, what proteomics has to tell
Abstract
Colorectal cancer is the third most common cancer and is highly fatal. During the last several years, research has been primarily based on the study of expression profiles using microarray technology. But now, investigators are putting into practice proteomic analyses of cancer tissues and cells to identify new diagnostic or therapeutic biomarkers for this cancer. Because the proteome reflects the state of a cell, tissue or organism more accurately, much is expected from proteomics to yield better tumor markers for disease diagnosis and therapy monitoring. This review summarizes the most relevant applications of proteomics the biomarker discovery for colorectal cancer.
Figures

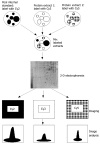
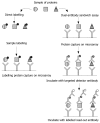
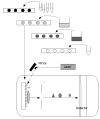
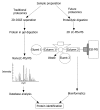
References
-
- Lièvre A, Laurent-Puig P. Molecular biology in clinical cancer research: the example of digestive cancers. Rev Epidemiol Sante Publique. 2005;53:267–282. - PubMed
-
- Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. - PubMed
-
- Penland SK, Goldberg RM. Current strategies in previously untreated advanced colorectal cancer. Oncology (Williston Park) 2004;18:715–722, 727; discussion 727-729. - PubMed
-
- Tanke HJ. Genomics and proteomics: the potential role of oral diagnostics. Ann N Y Acad Sci. 2007;1098:330–334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

