Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations
- PMID: 17991724
- PMCID: PMC2219306
- DOI: 10.1210/en.2007-1050
Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations
Abstract
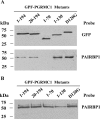
Progesterone (P4) receptor membrane component-1 (PGRMC1) and its binding partner, plasminogen activator inhibitor 1 RNA binding protein (PAIRBP1) are thought to form a complex that functions as membrane receptor for P4. The present investigations confirm PGRMC1's role in this membrane receptor complex by demonstrating that depleting PGMRC1 with PGRMC1 small interfering RNA results in a 60% decline in [(3)H]P4 binding and the loss of P4's antiapoptotic action. Studies conducted on partially purified GFP-PGRMC1 fusion protein indicate that [(3)H]P4 specifically binds to PGRMC1 at a single site with an apparent K(d) of about 35 nm. In addition, experiments using various deletion mutations reveal that the entire PGRMC1 molecule is required for maximal [(3)H]P4 binding and P4 responsiveness. Analysis of the binding data also suggests that the P4 binding site is within a segment of PGRMC1 that is composed of the transmembrane domain and the initial segment of the C terminus. Interestingly, PAIRBP1 appears to bind to the C terminus between amino acids 70-130, which is distal to the putative P4 binding site. Taken together, these data provide compelling evidence that PGRMC1 is the P4 binding protein that mediates P4's antiapoptotic action. Moreover, the deletion mutation studies indicate that each domain of PGRMC1 plays an essential role in modulating PGRMC1's capacity to both bind and respond to P4. Additional studies are required to more precisely delineate the role of each PGRMC1 domain in transducing P4's antiapoptotic action.
Figures








Similar articles
-
Plasminogen activator inhibitor 1 RNA-binding protein interacts with progesterone receptor membrane component 1 to regulate progesterone's ability to maintain the viability of spontaneously immortalized granulosa cells and rat granulosa cells.Biol Reprod. 2013 Jan 25;88(1):20. doi: 10.1095/biolreprod.112.103036. Print 2013 Jan. Biol Reprod. 2013. PMID: 23242527 Free PMC article.
-
Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action.Endocrinology. 2006 Jun;147(6):3133-40. doi: 10.1210/en.2006-0114. Epub 2006 Mar 2. Endocrinology. 2006. PMID: 16513825
-
Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression.Mol Cell Endocrinol. 2010 May 14;320(1-2):153-61. doi: 10.1016/j.mce.2010.02.005. Epub 2010 Feb 6. Mol Cell Endocrinol. 2010. PMID: 20144686 Free PMC article.
-
Progesterone signaling mediated through progesterone receptor membrane component-1 in ovarian cells with special emphasis on ovarian cancer.Steroids. 2011 Aug;76(9):903-9. doi: 10.1016/j.steroids.2011.02.011. Epub 2011 Mar 1. Steroids. 2011. PMID: 21371489 Free PMC article. Review.
-
Non-canonical progesterone signaling in granulosa cell function.Reproduction. 2014 Apr 8;147(5):R169-78. doi: 10.1530/REP-13-0582. Print 2014 May. Reproduction. 2014. PMID: 24516175 Free PMC article. Review.
Cited by
-
Cyclic Regulation of Sensory Perception by a Female Hormone Alters Behavior.Cell. 2015 Jun 4;161(6):1334-44. doi: 10.1016/j.cell.2015.04.052. Cell. 2015. PMID: 26046438 Free PMC article.
-
Plasminogen activator inhibitor 1 RNA-binding protein interacts with progesterone receptor membrane component 1 to regulate progesterone's ability to maintain the viability of spontaneously immortalized granulosa cells and rat granulosa cells.Biol Reprod. 2013 Jan 25;88(1):20. doi: 10.1095/biolreprod.112.103036. Print 2013 Jan. Biol Reprod. 2013. PMID: 23242527 Free PMC article.
-
Progesterone receptor membrane component 1 and its role in ovarian follicle growth.Front Neurosci. 2013 Jun 13;7:99. doi: 10.3389/fnins.2013.00099. eCollection 2013. Front Neurosci. 2013. PMID: 23781168 Free PMC article.
-
Progesterone Signaling and Mammalian Ovarian Follicle Growth Mediated by Progesterone Receptor Membrane Component Family Members.Cells. 2022 May 13;11(10):1632. doi: 10.3390/cells11101632. Cells. 2022. PMID: 35626669 Free PMC article. Review.
-
Epitope mapping of anti-PGRMC1 antibodies reveals the non-conventional membrane topology of PGRMC1 on the cell surface.Sci Rep. 2019 Jan 24;9(1):653. doi: 10.1038/s41598-018-37441-6. Sci Rep. 2019. PMID: 30679694 Free PMC article.
References
-
- Bramley T 2003 Non-genomic progesterone receptors in the mammalian ovary: some unresolved issues. Reproduction 125:3–15 - PubMed
-
- Peluso JJ 2006 Multiplicity of progesterone’s actions and receptors in the mammalian ovary. Biol Reprod 75:2–8 - PubMed
-
- Peluso JJ 2007 Non-genomic actions of progesterone in the normal and neoplastic mammalian ovary. Semin Reprod Med 25:198–207 - PubMed
-
- Rung E, Friberg PA, Shao R, Larsson DG, Nielsen E, Svensson PA, Carlsson B, Carlsson LM, Billig H 2005 Progesterone-receptor antagonists and statins decrease de novo cholesterol synthesis and increase apoptosis in rat and human periovulatory granulosa cells in vitro. Biol Reprod 72:538–545 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

