Premelanosome amyloid-like fibrils are composed of only golgi-processed forms of Pmel17 that have been proteolytically processed in endosomes
- PMID: 17991747
- PMCID: PMC2430631
- DOI: 10.1074/jbc.M708007200
Premelanosome amyloid-like fibrils are composed of only golgi-processed forms of Pmel17 that have been proteolytically processed in endosomes
Abstract
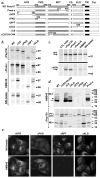
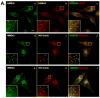
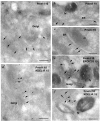
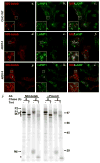
Melanin pigments are synthesized within specialized organelles called melanosomes and polymerize on intraluminal fibrils that form within melanosome precursors. The fibrils consist of proteolytic fragments derived from Pmel17, a pigment cell-specific integral membrane protein. The intracellular pathways by which Pmel17 accesses melanosome precursors and the identity of the Pmel17 derivatives within fibrillar melanosomes have been a matter of debate. We show here that antibodies that detect Pmel17 within fibrillar melanosomes recognize only the luminal products of proprotein convertase cleavage and not the remaining products linked to the transmembrane domain. Moreover, antibodies to the N and C termini detect only Pmel17 isoforms present in early biosynthetic compartments, which constitute a large fraction of detectable steady state Pmel17 in cell lysates because of slow early biosynthetic transport and rapid consumption by fibril formation. Using an antibody to a luminal epitope that is destroyed upon modification by O-linked oligosaccharides, we show that all post-endoplasmic reticulum Pmel17 isoforms are modified by Golgi-associated oligosaccharide transferases, and that only processed forms contribute to melanosome biogenesis. These data indicate that Pmel17 follows a single biosynthetic route from the endoplasmic reticulum through the Golgi complex and endosomes to melanosomes, and that only fragments encompassing previously described functional luminal determinants are present within the fibrils. These data have important implications for the site and mechanism of fibril formation.
Figures







Similar articles
-
Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction.J Biol Chem. 2010 May 21;285(21):16166-83. doi: 10.1074/jbc.M109.097725. Epub 2010 Mar 15. J Biol Chem. 2010. PMID: 20231267 Free PMC article.
-
N-terminal domains elicit formation of functional Pmel17 amyloid fibrils.J Biol Chem. 2009 Dec 18;284(51):35543-55. doi: 10.1074/jbc.M109.047449. J Biol Chem. 2009. PMID: 19840945 Free PMC article.
-
The Kringle-like Domain Facilitates Post-endoplasmic Reticulum Changes to Premelanosome Protein (PMEL) Oligomerization and Disulfide Bond Configuration and Promotes Amyloid Formation.J Biol Chem. 2016 Feb 12;291(7):3595-612. doi: 10.1074/jbc.M115.692442. Epub 2015 Dec 22. J Biol Chem. 2016. PMID: 26694611 Free PMC article.
-
PMEL Amyloid Fibril Formation: The Bright Steps of Pigmentation.Int J Mol Sci. 2016 Aug 31;17(9):1438. doi: 10.3390/ijms17091438. Int J Mol Sci. 2016. PMID: 27589732 Free PMC article. Review.
-
The Silver locus product Pmel17/gp100/Silv/ME20: controversial in name and in function.Pigment Cell Res. 2005 Oct;18(5):322-36. doi: 10.1111/j.1600-0749.2005.00269.x. Pigment Cell Res. 2005. PMID: 16162173 Free PMC article. Review.
Cited by
-
Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction.J Biol Chem. 2010 May 21;285(21):16166-83. doi: 10.1074/jbc.M109.097725. Epub 2010 Mar 15. J Biol Chem. 2010. PMID: 20231267 Free PMC article.
-
Research Techniques Made Simple: Cell Biology Methods for the Analysis of Pigmentation.J Invest Dermatol. 2020 Feb;140(2):257-268.e8. doi: 10.1016/j.jid.2019.12.002. J Invest Dermatol. 2020. PMID: 31980058 Free PMC article. Review.
-
Functional Domains and Evolutionary History of the PMEL and GPNMB Family Proteins.Molecules. 2021 Jun 9;26(12):3529. doi: 10.3390/molecules26123529. Molecules. 2021. PMID: 34207849 Free PMC article.
-
Amyloid form of ovalbumin evokes native antigen-specific immune response in the host: prospective immuno-prophylactic potential.J Biol Chem. 2015 Feb 13;290(7):4131-48. doi: 10.1074/jbc.M113.540989. Epub 2014 Dec 15. J Biol Chem. 2015. PMID: 25512377 Free PMC article.
-
A β-solenoid model of the Pmel17 repeat domain: insights to the formation of functional amyloid fibrils.J Comput Aided Mol Des. 2016 Feb;30(2):153-64. doi: 10.1007/s10822-015-9892-x. Epub 2016 Jan 11. J Comput Aided Mol Des. 2016. PMID: 26754844
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

