Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain
- PMID: 17991865
- PMCID: PMC4039561
- DOI: 10.1126/science.1147851
Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain
Abstract
The identification of neural stem and progenitor cells (NPCs) by in vivo brain imaging could have important implications for diagnostic, prognostic, and therapeutic purposes. We describe a metabolic biomarker for the detection and quantification of NPCs in the human brain in vivo. We used proton nuclear magnetic resonance spectroscopy to identify and characterize a biomarker in which NPCs are enriched and demonstrated its use as a reference for monitoring neurogenesis. To detect low concentrations of NPCs in vivo, we developed a signal processing method that enabled the use of magnetic resonance spectroscopy for the analysis of the NPC biomarker in both the rodent brain and the hippocampus of live humans. Our findings thus open the possibility of investigating the role of NPCs and neurogenesis in a wide variety of human brain disorders.
Figures
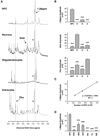
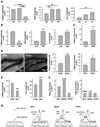
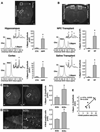
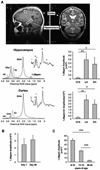
Comment in
-
Neuroscience. Spying on new neurons in the human brain.Science. 2007 Nov 9;318(5852):899-900. doi: 10.1126/science.318.5852.899a. Science. 2007. PMID: 17991833 No abstract available.
-
Comment on "magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain".Science. 2008 Aug 1;321(5889):640. doi: 10.1126/science.1153058. Science. 2008. PMID: 18669844 Free PMC article.
-
Comment on "Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain".Science. 2008 Aug 1;321(5889):640. doi: 10.1126/science.1153484. Science. 2008. PMID: 18669845 Free PMC article.
-
Comment on "Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain".Science. 2008 Aug 1;321(5889):640. doi: 10.1126/science.1153997. Science. 2008. PMID: 18669846
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

