Kinase packing defects as drug targets
- PMID: 17993409
- PMCID: PMC2136416
- DOI: 10.1016/j.drudis.2007.09.009
Kinase packing defects as drug targets
Erratum in
- Drug Discov Today. 2014 Apr;19(4):520
Abstract
Protein kinases constitute major targets in molecular cancer therapy. The structural conservation of kinases causes specificity problems in most drug inhibitors, often resulting in dangerous side effects. Here we survey recent approaches in drug design that exploit a molecular marker for specificity: the pattern of packing defects. These packing defects are solvent-exposed intramolecular hydrogen bonds that may be protected by drugs upon association. In this light, we review design strategies to achieve paralogue discrimination, to control cross reactivity and to overcome drug resistance induced by target mutations.
Figures
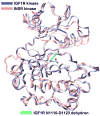

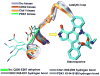



Similar articles
-
Anticipating drug resistance in the MAP kinase pathway.Pigment Cell Melanoma Res. 2010 Feb;23(1):7-9. doi: 10.1111/j.1755-148X.2009.00657.x. Epub 2009 Dec 2. Pigment Cell Melanoma Res. 2010. PMID: 19968818 Free PMC article. No abstract available.
-
Synergistic interactions between imatinib mesylate and the novel phosphoinositide-dependent kinase-1 inhibitor OSU-03012 in overcoming imatinib mesylate resistance.Blood. 2005 May 15;105(10):4021-7. doi: 10.1182/blood-2004-07-2967. Epub 2005 Jan 21. Blood. 2005. PMID: 15665113 Free PMC article.
-
Rational drug redesign to overcome drug resistance in cancer therapy: imatinib moving target.Cancer Res. 2007 May 1;67(9):4028-33. doi: 10.1158/0008-5472.CAN-07-0345. Cancer Res. 2007. PMID: 17483313 Free PMC article.
-
Mechanisms of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia and recent therapeutic strategies to overcome resistance.Hematology Am Soc Hematol Educ Program. 2009:461-76. doi: 10.1182/asheducation-2009.1.461. Hematology Am Soc Hematol Educ Program. 2009. PMID: 20008232 Review.
-
Imatinib-refractory gastrointestinal stromal tumors: the clinical problem and therapeutic strategies.Curr Oncol Rep. 2006 May;8(3):192-7. doi: 10.1007/s11912-006-0019-3. Curr Oncol Rep. 2006. PMID: 16618383 Review.
References
-
- Dancey J, Sausville EA. Issues and progress with protein kinase inhibitors for cancer treatment. Nat Rev Drug Discov. 2003;2:296–313. - PubMed
-
- Levitski A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;267:1782–1788. - PubMed
-
- Donato NJ, Talpaz M. Clinical use of tyrosine kinase inhibitors: Therapy for chronic myelogenous leukemia and other cancers. Clin Cancer Res. 2000;6:2965–2966. - PubMed
-
- Tibes R, et al. Tyrosine kinase inhibitors and the dawn of molecular cancer therapeutics. Annu Rev Pharmacol Toxicol. 2005;45:357–384. - PubMed
-
- Gibbs J, Oliff A. Pharmaceutical research in molecular oncology. Cell. 1994;79:193–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

