Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex
- PMID: 17993457
- PMCID: PMC2186065
- DOI: 10.1074/jbc.M707907200
Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex
Abstract
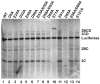
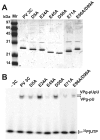
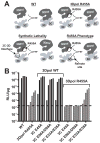
Picornaviruses have a peptide termed VPg covalently linked to the 5'-end of the genome. Attachment of VPg to the genome occurs in at least two steps. First, Tyr-3 of VPg, or some precursor thereof, is used as a primer by the viral RNA-dependent RNA polymerase, 3Dpol, to produce VPg-pUpU. Second, VPg-pUpU is used as a primer to produce full-length genomic RNA. Production of VPg-pUpU is templated by a single adenylate residue located in the loop of an RNA stem-loop structure termed oriI by using a slide-back mechanism. Recruitment of 3Dpol to and its stability on oriI have been suggested to require an interaction between the back of the thumb subdomain of 3Dpol and an undefined region of the 3C domain of viral protein 3CD. We have performed surface acidic-to-alanine-scanning mutagenesis of 3C to identify the surface of 3C with which 3Dpol interacts. This analysis identified numerous viable poliovirus mutants with reduced growth kinetics that correlated to reduced kinetics of RNA synthesis that was attributable to a change in VPg-pUpU production. Importantly, these 3C derivatives were all capable of binding to oriI as well as wild-type 3C. Synthetic lethality was observed for these mutants when placed in the context of a poliovirus mutant containing 3Dpol-R455A, a residue on the back of the thumb required for VPg uridylylation. These data were used to guide molecular docking of the structures for a poliovirus 3C dimer and 3Dpol, leading to a structural model for the 3C(2)-3Dpol complex that extrapolates well to all picornaviruses.
Figures
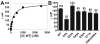


Similar articles
-
Human rhinovirus type 14 gain-of-function mutants for oriI utilization define residues of 3C(D) and 3Dpol that contribute to assembly and stability of the picornavirus VPg uridylylation complex.J Virol. 2007 Nov;81(22):12485-95. doi: 10.1128/JVI.00972-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855535 Free PMC article.
-
Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex.J Biol Chem. 2007 Jun 1;282(22):16202-13. doi: 10.1074/jbc.M610608200. Epub 2007 Mar 27. J Biol Chem. 2007. PMID: 17392285 Free PMC article.
-
Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation.J Biol Chem. 2002 Aug 30;277(35):31551-62. doi: 10.1074/jbc.M204408200. Epub 2002 Jun 19. J Biol Chem. 2002. PMID: 12077141
-
Formation and working mechanism of the picornavirus VPg uridylylation complex.Curr Opin Virol. 2014 Dec;9:24-30. doi: 10.1016/j.coviro.2014.09.003. Epub 2014 Sep 19. Curr Opin Virol. 2014. PMID: 25240314 Review.
-
Uridylylation of the genome-linked protein of poliovirus in vitro is dependent upon an endogenous RNA template.Virus Res. 1987 Sep;8(3):193-204. doi: 10.1016/0168-1702(87)90015-3. Virus Res. 1987. PMID: 2825442 Review.
Cited by
-
Picornavirus RNA polyadenylation by 3D(pol), the viral RNA-dependent RNA polymerase.Virus Res. 2015 Aug 3;206:3-11. doi: 10.1016/j.virusres.2014.12.030. Epub 2015 Jan 3. Virus Res. 2015. PMID: 25559071 Free PMC article. Review.
-
Interaction of Sesbania mosaic virus (SeMV) RNA-dependent RNA polymerase (RdRp) with the p10 domain of polyprotein 2a and its implications in SeMV replication.FEBS Open Bio. 2014 Mar 29;4:362-9. doi: 10.1016/j.fob.2014.03.009. eCollection 2014. FEBS Open Bio. 2014. PMID: 24918050 Free PMC article.
-
Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants.J Virol. 2009 Sep;83(18):9370-87. doi: 10.1128/JVI.02076-08. Epub 2009 Jul 8. J Virol. 2009. PMID: 19587035 Free PMC article.
-
UGGT1 enhances enterovirus 71 pathogenicity by promoting viral RNA synthesis and viral replication.PLoS Pathog. 2017 May 17;13(5):e1006375. doi: 10.1371/journal.ppat.1006375. eCollection 2017 May. PLoS Pathog. 2017. PMID: 28545059 Free PMC article.
-
Expanding knowledge of P3 proteins in the poliovirus lifecycle.Future Microbiol. 2010 Jun;5(6):867-81. doi: 10.2217/fmb.10.40. Future Microbiol. 2010. PMID: 20521933 Free PMC article. Review.
References
-
- Racaniello VR. Picornaviridae: The viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 4. Lippincott-Raven Publishers; Philadelphia, PA: 2001.
-
- Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. ASM Press; Washington, D.C.: 2002.
-
- Paul AV. Possible Unifying Mechanism of Picornavirus Genome Replication. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. 1. ASM Press; Washington, D.C: 2002.
-
- Paul AV, van Boom JH, Filippov D, Wimmer E. Nature. 1998;393(6682):280–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

