Kinetics of multiplex hybridization: mechanisms and implications
- PMID: 17993490
- PMCID: PMC2242776
- DOI: 10.1529/biophysj.107.121459
Kinetics of multiplex hybridization: mechanisms and implications
Abstract
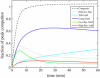
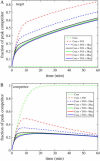
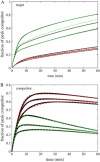
Quantitative analysis of DNA microarray data is complicated by uncertainties inherent to the experimental setup. Using computer simulations and real-time experimental results, we have previously demonstrated effects of multiplex reactions on a single sensing zone of an array, which may be a leading factor in erroneous interpretation of experimental data. We suggest here that a simplified three-component kinetic model may present a sufficient approximation to describe the general case of DNA sensing in a complex sample milieu. We show that, by analyzing the real-time hybridization kinetics of a nontarget species, we can perform quantitative analysis of unlabeled targets of interest within a broad dynamic range of concentrations.
Figures


Similar articles
-
Microarray results improve significantly as hybridization approaches equilibrium.Biotechniques. 2004 May;36(5):790-6. doi: 10.2144/04365ST02. Biotechniques. 2004. PMID: 15152598
-
Mass transfer effects on DNA hybridization in a flow-through microarray.J Biotechnol. 2009 Jan 15;139(2):179-85. doi: 10.1016/j.jbiotec.2008.10.001. Epub 2008 Oct 15. J Biotechnol. 2009. PMID: 18984014
-
The limits of log-ratios.BMC Biotechnol. 2004 Mar 8;4:3. doi: 10.1186/1472-6750-4-3. BMC Biotechnol. 2004. PMID: 15113428 Free PMC article.
-
Principles of gene microarray data analysis.Adv Exp Med Biol. 2007;593:19-30. doi: 10.1007/978-0-387-39978-2_3. Adv Exp Med Biol. 2007. PMID: 17265713 Review.
-
High-density arrays and insights into genome function.Biotechnol Genet Eng Rev. 2000;17:109-46. doi: 10.1080/02648725.2000.10647990. Biotechnol Genet Eng Rev. 2000. PMID: 11255664 Review. No abstract available.
Cited by
-
Direct DNA methylation profiling using methyl binding domain proteins.Anal Chem. 2010 Jun 15;82(12):5012-9. doi: 10.1021/ac1010316. Anal Chem. 2010. PMID: 20507169 Free PMC article.
-
Salt concentration effects on equilibrium melting curves from DNA microarrays.Biophys J. 2010 Sep 22;99(6):1886-95. doi: 10.1016/j.bpj.2010.07.002. Biophys J. 2010. PMID: 20858434 Free PMC article.
-
Optimization of encoded hydrogel particles for nucleic acid quantification.Anal Chem. 2009 Jun 15;81(12):4873-81. doi: 10.1021/ac9005292. Anal Chem. 2009. PMID: 19435332 Free PMC article.
-
Real-time DNA microarrays: reality check.Biochem Soc Trans. 2009 Apr;37(Pt 2):471-5. doi: 10.1042/BST0370471. Biochem Soc Trans. 2009. PMID: 19290884 Free PMC article. Review.
-
BIOPHYSICAL PROPERTIES OF NUCLEIC ACIDS AT SURFACES RELEVANT TO MICROARRAY PERFORMANCE.Biomater Sci. 2014 Apr 1;2(4):436-471. doi: 10.1039/C3BM60181A. Biomater Sci. 2014. PMID: 24765522 Free PMC article.
References
-
- Levicky R., Horgan A. Physicochemical perspectives on DNA microarray and biosensor technologies. Trends Biotechnol. 2005;23:143–149. - PubMed
-
- Provenzano M., Mocellin S. Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv. Exp. Med. Biol. 2007;593:66–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

