Microscopic simulation of membrane molecule diffusion on corralled membrane surfaces
- PMID: 17993499
- PMCID: PMC2242759
- DOI: 10.1529/biophysj.107.106484
Microscopic simulation of membrane molecule diffusion on corralled membrane surfaces
Abstract
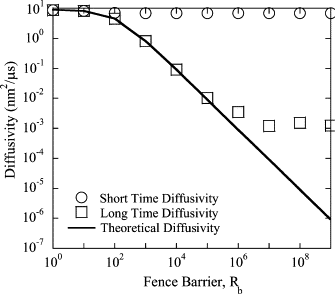
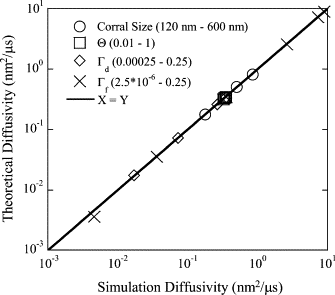
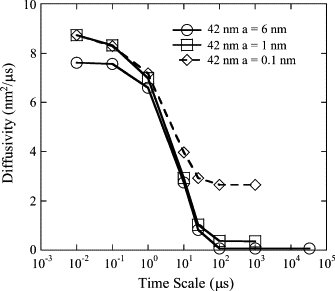
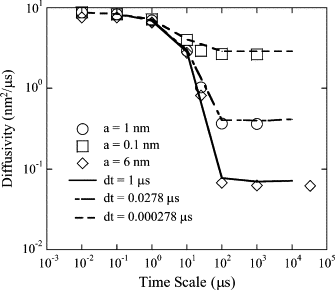
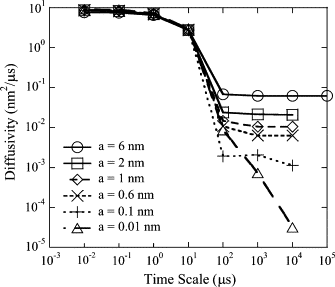
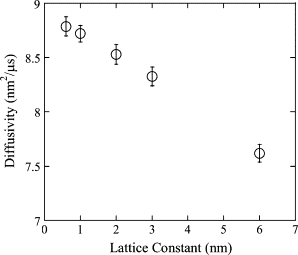
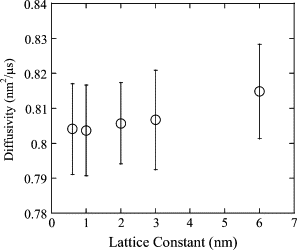
The current understanding of how receptors diffuse and cluster in the plasma membrane is limited. Data from single-particle tracking and laser tweezer experiments have suggested that membrane molecule diffusion is affected by the presence of barriers dividing the membrane into corrals. Here, we have developed a stochastic spatial model to simulate the effect of corrals on the diffusion of molecules in the plasma membrane. The results of this simulation confirm that a fence barrier (the ratio of the transition probability for diffusion across a boundary to that within a corral) on the order of 10(3)-10(4) recreates the experimentally measured difference in diffusivity between artificial and natural plasma membranes. An expression for the macroscopic diffusivity of receptors on corralled membranes is derived to analyze the effects of the corral parameters on diffusion rate. We also examine whether the lattice model is an appropriate description of the plasma membrane and look at three different sets of boundary conditions that describe diffusion over the barriers and whether diffusion events on the plasma membrane may occur with a physically relevant length scale. Finally, we show that to observe anomalous (two-timescale) diffusion, one needs high temporal (microsecond) resolution along with sufficiently long (more than milliseconds) trajectories.
Figures





References
-
- Johnson G.L., Vaillancourt R.R. Sequential protein kinase reactions controlling cell growth and differentiation. Curr. Opin. Cell Biol. 1994;6:230–238. - PubMed
-
- Saez-Rodrigues J., Kremling A., Conzelmann H., Bettenbrock K., Gilles E.D. Modular analysis of signal transduction networks. IEEE Contr. Syst. Mag. 2004;24:35–52.
-
- Holbro T., Civenni G., Hynes N.E. The ErbB receptors and their role in cancer progression. Exp. Cell Res. 2003;284:99–110. - PubMed
-
- Nicholson R.I., Gee J.M.W., Harper M.E. EGFR and cancer prognosis. Eur. J. Cancer. 2001;37:S9–S15. - PubMed
-
- Jorissen R.N., Walker F., Pouliot N., Garrett T.P.J., Ward C.W., Burgess A.W. Epidermal growth factor receptor: mechanisms of activation and signaling. Exp. Cell Res. 2003;284:31–53. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

