Transcriptome analysis of a phenol-producing Pseudomonas putida S12 construct: genetic and physiological basis for improved production
- PMID: 17993537
- PMCID: PMC2293262
- DOI: 10.1128/JB.01379-07
Transcriptome analysis of a phenol-producing Pseudomonas putida S12 construct: genetic and physiological basis for improved production
Abstract
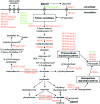
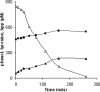
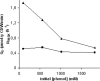
The unknown genetic basis for improved phenol production by a recombinant Pseudomonas putida S12 derivative bearing the tpl (tyrosine-phenol lyase) gene was investigated via comparative transcriptomics, nucleotide sequence analysis, and targeted gene disruption. We show upregulation of tyrosine biosynthetic genes and possibly decreased biosynthesis of tryptophan caused by a mutation in the trpE gene as the genetic basis for the enhanced phenol production. In addition, several genes in degradation routes connected to the tyrosine biosynthetic pathway were upregulated. This either may be a side effect that negatively affects phenol production or may point to intracellular accumulation of tyrosine or its intermediates. A number of genes identified by the transcriptome analysis were selected for targeted disruption in P. putida S12TPL3. Physiological and biochemical examination of P. putida S12TPL3 and these mutants led to the conclusion that the metabolic flux toward tyrosine in P. putida S12TPL3 was improved to such an extent that the heterologous tyrosine-phenol lyase enzyme had become the rate-limiting step in phenol biosynthesis.
Figures


References
-
- Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernandez, B. Galan, J. L. Garcia, E. Diaz, and B. Minambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 1865062-5077. - PMC - PubMed
-
- Ballerstedt, H., R. J. M. Volkers, A. E. Mars, J. E. Hallsworth, V. A. Martins dos Santos, J. Puchalka, J. Duuren, G. Eggink, K. N. Timmis, J. A. M. de Bont, and J. Wery. 2007. Genomotyping of Pseudomonas putida strains using P. putida KT2440-based high-density DNA microarrays: implications for transcriptomics studies. Appl. Microbiol. Biotechnol. 751133-1142. - PMC - PubMed
-
- Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 1471611-1620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

