Bifidobacteria in feces and environmental waters
- PMID: 17993557
- PMCID: PMC2227705
- DOI: 10.1128/AEM.01221-07
Bifidobacteria in feces and environmental waters
Abstract
Bifidobacteria have been recommended as potential indicators of human fecal pollution in surface waters even though very little is known about their presence in nonhuman fecal sources. The objective of this research was to shed light on the occurrence and molecular diversity of this fecal indicator group in different animals and environmental waters. Genus- and species-specific 16S rRNA gene PCR assays were used to study the presence of bifidobacteria among 269 fecal DNA extracts from 32 different animals. Twelve samples from three wastewater treatment plants and 34 water samples from two fecally impacted watersheds were also tested. The species-specific assays showed that Bifidobacterium adolescentis, B. bifidum, B. dentium, and B. catenulatum had the broadest host distribution (11.9 to 17.4%), whereas B. breve, B. infantis, and B. longum were detected in fewer than 3% of all fecal samples. Phylogenetic analysis of 356 bifidobacterial clones obtained from different animal feces showed that ca. 67% of all of the sequences clustered with cultured bifidobacteria, while the rest formed a supercluster with low sequence identity (i.e., <94%) to previously described Bifidobacterium spp. The B. pseudolongum subcluster (>97% similarity) contained 53 fecal sequences from seven different animal hosts, suggesting the cosmopolitan distribution of members of this clade. In contrast, two clades containing B. thermophilum and B. boum clustered exclusively with 37 and 18 pig fecal clones, respectively, suggesting host specificity. Using species-specific assays, bifidobacteria were detected in only two of the surface water DNA extracts, although other fecal anaerobic bacteria were detected in these waters. Overall, the results suggest that the use of bifidobacterial species as potential markers to monitor human fecal pollution in natural waters may be questionable.
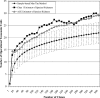
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- American Public Health Association. 1998. Standard methods for the examination of water and wastewater. APHA/AWWA/WEF, Washington, DC.
-
- Biavati, B., V. Scardovi, and W. E. C. Moore. 1982. Electrophoretic patterns of proteins in the genus Bifidobacterium and proposal of four new species. Int. J. Syst. Bacteriol. 32:358-373.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

