Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae
- PMID: 17993564
- PMCID: PMC2223240
- DOI: 10.1128/AEM.01531-07
Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae
Abstract
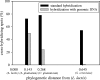
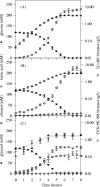
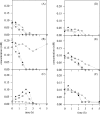
The study of microbial interactions in mixed cultures remains an important conceptual and methodological challenge for which transcriptome analysis could prove to be the essential method for improving our understanding. However, the use of whole-genome DNA chips is often restricted to the pure culture of the species for which the chips were designed. In this study, massive cross-hybridization was observed between the foreign cDNA and the specific Lactococcus lactis DNA chip. A very simple method is proposed to considerably decrease this nonspecific hybridization, consisting of adding the microbial partner's DNA. A correlation was established between the resulting cross-hybridization and the phylogenetic distance between the microbial partners. The response of L. lactis to the presence of Saccharomyces cerevisiae was analyzed during the exponential growth phase in fermentors under defined growth conditions. Although no differences between growth kinetics were observed for the pure and the mixed cultures of L. lactis, the mRNA levels of 158 genes were significantly modified. More particularly, a strong reorientation of pyrimidine metabolism was observed when L. lactis was grown in mixed cultures. These changes in transcript abundance were demonstrated to be regulated by the ethanol produced by the yeast and were confirmed by an independent method (quantitative reverse transcription-PCR).
Figures

References
-
- Arsene-Ploetze, F., V. Kugler, J. Martinussen, and F. Bringel. 2006. Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1. J. Bacteriol. 188:8607-8616. - PMC - PubMed
-
- Barrière, C., M. Veiga-da-Cunha, N. Pons, E. Guédon, S. A. van Hijum, J. Kok, O. P. Kuipers, D. S. Ehrlich, and P. Renault. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752-3761. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

