ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis
- PMID: 17993621
- PMCID: PMC2174867
- DOI: 10.1105/tpc.107.054494
ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis
Abstract
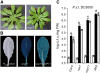
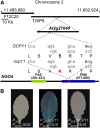
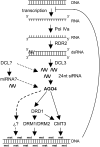
Here, we report the characterization of the Arabidopsis thaliana ocp11 (for overexpressor of cationic peroxidase11) mutant, in which a beta-glucuronidase reporter gene under the control of the H(2)O(2)-responsive Ep5C promoter is constitutively expressed. ocp11 plants show enhanced disease susceptibility to the virulent bacterium Pseudomonas syringae pv tomato DC3000 (P.s.t. DC3000) and also to the avirulent P.s.t. DC3000 carrying the effector avrRpm1 gene. In addition, ocp11 plants are also compromised in resistance to the nonhost pathogen P. syringae pv tabaci. Genetic and molecular analyses reveal that ocp11 plants are not affected in salicylic acid perception. We cloned OCP11 and show that it encodes ARGONAUTE4 (AGO4), a component of the pathway that mediates the transcriptional gene silencing associated with small interfering RNAs that direct DNA methylation at specific loci, a phenomenon known as RNA-directed DNA methylation (RdDM). Thus, we renamed our ocp11 mutant ago4-2, as it represents a different allele to the previously characterized recessive ago4-1. Both mutants decrease the extent of DNA cytosine methylation at CpNpG and CpHpH (asymmetric) positions present at different DNA loci and show commonalities in all of the molecular and phenotypic aspects that we have considered. Interestingly, we show that AGO4 works independently of other components of the RdDM pathway in mediating resistance to P.s.t. DC3000, and loss of function in other components of the pathway operating upstream of AGO4, such as RDR2 and DCL3, or operating downstream, such as DRD1, CMT3, DRM1, and DRM2, does not compromise resistance to this pathogen.
Figures
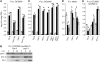

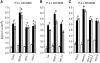

References
-
- Abramovitch, R.B., and Martin, G.B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7 356–364. - PubMed
-
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. - PubMed
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

