Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs
- PMID: 17993623
- PMCID: PMC2174889
- DOI: 10.1105/tpc.107.053645
Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs
Abstract
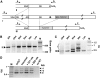
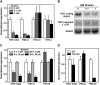
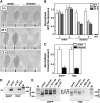
The most widespread riboswitch class, found in organisms from all three domains of life, is responsive to the vitamin B(1) derivative thiamin pyrophosphate (TPP). We have established that a TPP-sensing riboswitch is present in the 3' untranslated region (UTR) of the thiamin biosynthetic gene THIC of all plant species examined. The THIC TPP riboswitch controls the formation of transcripts with alternative 3' UTR lengths, which affect mRNA accumulation and protein production. We demonstrate that riboswitch-mediated regulation of alternative 3' end processing is critical for TPP-dependent feedback control of THIC expression. Our data reveal a mechanism whereby metabolite-dependent alteration of RNA folding controls splicing and alternative 3' end processing of mRNAs. These findings highlight the importance of metabolite sensing by riboswitches in plants and further reveal the significance of alternative 3' end processing as a mechanism of gene control in eukaryotes.
Figures

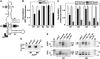


References
-
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., Wickiser, J.K., and Breaker, R.R. (2004). New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 101 6421–6426. - PMC - PubMed
-
- Batey, R.T., Gilbert, S.D., and Montange, R.K. (2004). Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432 411–415. - PubMed
-
- Cazzonelli, C.I., and Velten, J. (2006). An in vivo, luciferase-based, Agrobacterium-infiltration assay system: Implications for post-transcriptional gene silencing. Planta 224 582–597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

