Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia
- PMID: 17993624
- PMCID: PMC2174892
- DOI: 10.1105/tpc.107.053322
Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia
Abstract
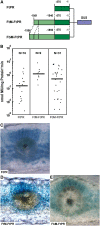
C(4) photosynthesis presents a sophisticated integration of two complementary cell types, mesophyll and bundle sheath cells. It relies on the differential expression of the genes encoding the component enzymes and transporters of this pathway. The entry enzyme of C(4) photosynthesis, phosphoenolpyruvate carboxylase (PEPC), is found exclusively in mesophyll cells, and the expression of the corresponding gene is regulated at the transcriptional level. In the C(4) dicot Flaveria trinervia, the mesophyll-specific expression of the C(4) PEPC gene (ppcA) depends on a 41-bp segment in the distal promoter region referred to as MEM1 (for mesophyll expression module1). Here, we show that a MEM1 sequence found in the orthologous ppcA gene from the C(3) species Flaveria pringlei is not able to direct mesophyll-specific gene expression. The two orthologous MEM1 sequences of F. pringlei and F. trinervia differ at two positions, a G-to-A exchange and the insertion of the tetranucleotide CACT. Changes at these two positions in the C(3) MEM1 sequence were necessary and sufficient to create a mesophyll-specificity element during C(4) evolution. The MEM1 of F. trinervia enhances mesophyll expression and concomitantly represses expression in bundle sheath cells and vascular bundles.
Figures




Similar articles
-
cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene.Plant Cell. 2004 May;16(5):1077-90. doi: 10.1105/tpc.019729. Epub 2004 Apr 20. Plant Cell. 2004. PMID: 15100398 Free PMC article.
-
A MEM1-like motif directs mesophyll cell-specific expression of the gene encoding the C4 carbonic anhydrase in Flaveria.J Exp Bot. 2017 Jan;68(2):311-320. doi: 10.1093/jxb/erw475. Epub 2016 Dec 30. J Exp Bot. 2017. PMID: 28040798 Free PMC article.
-
Evolution of c4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria.Ann Bot. 2004 Jan;93(1):13-23. doi: 10.1093/aob/mch003. Epub 2003 Nov 26. Ann Bot. 2004. PMID: 14644912 Free PMC article. Review.
-
Evolution of the C4 phosphoenolpyruvate carboxylase promoter of the C4 species Flaveria trinervia: the role of the proximal promoter region.BMC Plant Biol. 2008 Jan 21;8:4. doi: 10.1186/1471-2229-8-4. BMC Plant Biol. 2008. PMID: 18208593 Free PMC article.
-
Evolution of C4 phosphoenolpyruvate carboxylase.Arch Biochem Biophys. 2003 Jun 15;414(2):180-8. doi: 10.1016/s0003-9861(03)00165-6. Arch Biochem Biophys. 2003. PMID: 12781769 Review.
Cited by
-
Identification of Leaf Promoters for Use in Transgenic Wheat.Plants (Basel). 2018 Mar 28;7(2):27. doi: 10.3390/plants7020027. Plants (Basel). 2018. PMID: 29597282 Free PMC article.
-
C4 cycles: past, present, and future research on C4 photosynthesis.Plant Cell. 2011 Nov;23(11):3879-92. doi: 10.1105/tpc.111.092098. Epub 2011 Nov 29. Plant Cell. 2011. PMID: 22128120 Free PMC article. Review.
-
OBV (obscure vein), a C2H2 zinc finger transcription factor, positively regulates chloroplast development and bundle sheath extension formation in tomato (Solanum lycopersicum) leaf veins.Hortic Res. 2021 Nov 1;8(1):230. doi: 10.1038/s41438-021-00659-z. Hortic Res. 2021. PMID: 34719693 Free PMC article.
-
Challenges and Approaches to Crop Improvement Through C3-to-C4 Engineering.Front Plant Sci. 2021 Sep 14;12:715391. doi: 10.3389/fpls.2021.715391. eCollection 2021. Front Plant Sci. 2021. PMID: 34594351 Free PMC article. Review.
-
Most of the tight positional conservation of transcription factor binding sites near the transcription start site reflects their co-localization within regulatory modules.BMC Bioinformatics. 2016 Nov 21;17(1):479. doi: 10.1186/s12859-016-1354-5. BMC Bioinformatics. 2016. PMID: 27871221 Free PMC article.
References
-
- Akyildiz, M. (2007). Identification of cis- and trans-Regulatory Factors Controlling the Expression of the C4 Phosphoenolpyruvate Carboxylase Gene of the C4 Dicot Flaveria trinervia. PhD dissertation (Düsseldorf, Germany: Heinrich-Heine Universität).
-
- Blackwood, E.M., and Kadonaga, J.T. (1998). Going the distance: A current view of enhancer action. Science 281 60–63. - PubMed
-
- Bläsing, O.E., Ernst, K., Streubel, M., Westhoff, P., and Svensson, P. (2002). The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia—Implications for the evolution of C4 photosynthesis. Planta 215 448–456. - PubMed
-
- Brown, N.J., Parsley, K., and Hibberd, J.M. (2005). The future of C4 research—maize, Flaveria or Cleome? Trends Plant Sci. 10 215–221. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

