RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo
- PMID: 17994019
- PMCID: PMC2824921
- DOI: 10.1038/ng.2007.26
RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo
Abstract
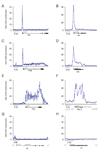
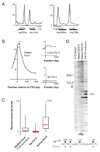
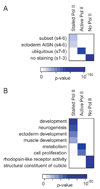
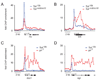
It is widely assumed that the key rate-limiting step in gene activation is the recruitment of RNA polymerase II (Pol II) to the core promoter. Although there are well-documented examples in which Pol II is recruited to a gene but stalls, a general role for Pol II stalling in development has not been established. We have carried out comprehensive Pol II chromatin immunoprecipitation microarray (ChIP-chip) assays in Drosophila embryos and identified three distinct Pol II binding behaviors: active (uniform binding across the entire transcription unit), no binding, and stalled (binding at the transcription start site). The notable feature of the approximately 10% genes that are stalled is that they are highly enriched for developmental control genes, which are either repressed or poised for activation during later stages of embryogenesis. We propose that Pol II stalling facilitates rapid temporal and spatial changes in gene activity during development.
Figures


Comment in
-
Stalled polymerases and transcriptional regulation.Nat Genet. 2007 Dec;39(12):1421-2. doi: 10.1038/ng1207-1421. Nat Genet. 2007. PMID: 18046324 No abstract available.
References
-
- Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem Sci. 2005;30:275–279. - PubMed
-
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. - PubMed
-
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

