RNA polymerase is poised for activation across the genome
- PMID: 17994021
- PMCID: PMC2365887
- DOI: 10.1038/ng.2007.21
RNA polymerase is poised for activation across the genome
Abstract
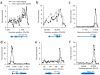
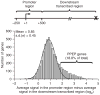
Regulation of gene expression is integral to the development and survival of all organisms. Transcription begins with the assembly of a pre-initiation complex at the gene promoter, followed by initiation of RNA synthesis and the transition to productive elongation. In many cases, recruitment of RNA polymerase II (Pol II) to a promoter is necessary and sufficient for activation of genes. However, there are a few notable exceptions to this paradigm, including heat shock genes and several proto-oncogenes, whose expression is attenuated by regulated stalling of polymerase elongation within the promoter-proximal region. To determine the importance of polymerase stalling for transcription regulation, we carried out a genome-wide search for Drosophila melanogaster genes with Pol II stalled within the promoter-proximal region. Our data show that stalling is widespread, occurring at hundreds of genes that respond to stimuli and developmental signals. This finding indicates a role for regulation of polymerase elongation in the transcriptional responses to dynamic environmental and developmental cues.
Figures

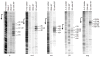
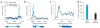

Comment in
-
Stalled polymerases and transcriptional regulation.Nat Genet. 2007 Dec;39(12):1421-2. doi: 10.1038/ng1207-1421. Nat Genet. 2007. PMID: 18046324 No abstract available.
References
-
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. - PubMed
-
- Luse DS, Samkurashvili I. The transition from initiation to elongation by RNA polymerase II. Cold Spring Harb Symp Quant Biol. 1998;63:289–300. - PubMed
-
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. - PubMed
-
- Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

