Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures
- PMID: 17994088
- PMCID: PMC2474711
- DOI: 10.1038/nature06340
Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures
Abstract
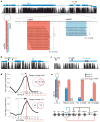
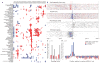
Sequencing of multiple related species followed by comparative genomics analysis constitutes a powerful approach for the systematic understanding of any genome. Here, we use the genomes of 12 Drosophila species for the de novo discovery of functional elements in the fly. Each type of functional element shows characteristic patterns of change, or 'evolutionary signatures', dictated by its precise selective constraints. Such signatures enable recognition of new protein-coding genes and exons, spurious and incorrect gene annotations, and numerous unusual gene structures, including abundant stop-codon readthrough. Similarly, we predict non-protein-coding RNA genes and structures, and new microRNA (miRNA) genes. We provide evidence of miRNA processing and functionality from both hairpin arms and both DNA strands. We identify several classes of pre- and post-transcriptional regulatory motifs, and predict individual motif instances with high confidence. We also study how discovery power scales with the divergence and number of species compared, and we provide general guidelines for comparative studies.
Figures




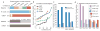
Comment in
-
Evolutionary genomics: come fly with us.Nature. 2007 Nov 8;450(7167):184-5. doi: 10.1038/450184a. Nature. 2007. PMID: 17994082 No abstract available.
References
-
- Miller W, Makova KD, Nekrutenko A, Hardison RC. Comparative genomics. Annu Rev Genomics Hum Genet. 2004;5:15–56. - PubMed
-
- Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nature Rev Genet. 2003;4:251–262. - PubMed
-
- Kellis M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. - PubMed
-
- Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. - PubMed
-
- Brent MR. Genome annotation past, present, and future: how to define an ORF at each locus. Genome Res. 2005;15:1777–1786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

