Multiplexed detection methods for profiling microRNA expression in biological samples
- PMID: 17994653
- PMCID: PMC2739110
- DOI: 10.1002/anie.200702450
Multiplexed detection methods for profiling microRNA expression in biological samples
Abstract
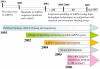
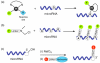
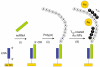
The recent discovery of short, non-protein coding RNA molecules, such as microRNA molecules (miRNAs), that can control gene expression has unveiled a whole new layer of complexity in the regulation of cell function. Since 2001, there has been a surge of interest in understanding the regulatory role of the hundreds to thousands of miRNAs expressed in both plants and animals. Significant progress in this area requires the development of quantitative bioanalytical methods for the rapid, multiplexed detection of all miRNAs that are present in a particular cell or tissue sample. In this Minireview, we discuss some of the latest methods for high-throughput miRNA profiling and the unique technological challenges that must be surmounted in this endeavor.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

