Self-assembly of human latexin into amyloid-like oligomers
- PMID: 17996039
- PMCID: PMC2212644
- DOI: 10.1186/1472-6807-7-75
Self-assembly of human latexin into amyloid-like oligomers
Abstract
Background: In conformational disorders, it is not evident which amyloid aggregates affect specific molecular mechanisms or cellular pathways, which cause disease because of their quantity and mechanical features and which states in aggregate formation are pathogenic. Due to the increasing consensus that prefibrillar oligomers play a major role in conformational diseases, there is a growing interest in understanding the characteristics of metastable polypeptide associations.
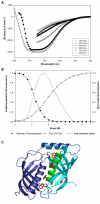
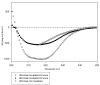
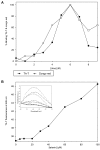
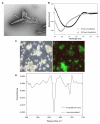
Results: Here, we show that human latexin, a protein that shares the same fold with cystatin C, assembles into stable spherical amyloid-like oligomers that bind thioflavin-T and congo red similarly to common amyloid structures but do not evolve into fibrils. Latexin self-assembly correlates with the formation of a mostly denaturated state rather than with the population of partially structured intermediates during the unfolding process. The results suggest that unfolding of alpha-helix 3 might be involved in the transition of latexin toward amyloidotic species, supporting the notion of the protective role of the native protein structure against polymerization.
Conclusion: Overall the data herein indicate that latexin could be a good model for the study of the structural and sequential determinants of oligomeric assemblies in protein aggregation processes.
Figures



Similar articles
-
Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure.Biochem J. 2012 May 1;443(3):719-26. doi: 10.1042/BJ20111924. Biochem J. 2012. PMID: 22316405
-
Kinetically driven refolding of the hyperstable EBNA1 origin DNA-binding dimeric beta-barrel domain into amyloid-like spherical oligomers.Proteins. 2008 Feb 1;70(2):450-61. doi: 10.1002/prot.21580. Proteins. 2008. PMID: 17680697
-
Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy.Biophys J. 2005 Jun;88(6):4200-12. doi: 10.1529/biophysj.104.049700. Epub 2005 Mar 11. Biophys J. 2005. PMID: 15764666 Free PMC article.
-
Cytotoxic species in amyloid-associated diseases: Oligomers or mature fibrils.Adv Protein Chem Struct Biol. 2019;118:333-369. doi: 10.1016/bs.apcsb.2019.06.001. Epub 2019 Aug 7. Adv Protein Chem Struct Biol. 2019. PMID: 31928731 Review.
-
Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
Cited by
-
Aging stem cells, latexin, and longevity.Exp Cell Res. 2008 Jun 10;314(9):1962-72. doi: 10.1016/j.yexcr.2008.01.032. Epub 2008 Feb 19. Exp Cell Res. 2008. PMID: 18374916 Free PMC article. Review. No abstract available.
-
Multiple oxygen tension environments reveal diverse patterns of transcriptional regulation in primary astrocytes.PLoS One. 2011;6(6):e21638. doi: 10.1371/journal.pone.0021638. Epub 2011 Jun 27. PLoS One. 2011. PMID: 21738745 Free PMC article.
-
Large Gliadin Peptides Detected in the Pancreas of NOD and Healthy Mice following Oral Administration.J Diabetes Res. 2016;2016:2424306. doi: 10.1155/2016/2424306. Epub 2016 Oct 4. J Diabetes Res. 2016. PMID: 27795959 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

