Global patterns of sequence evolution in Drosophila
- PMID: 17996078
- PMCID: PMC2180185
- DOI: 10.1186/1471-2164-8-408
Global patterns of sequence evolution in Drosophila
Abstract
Background: Sequencing of the genomes of several Drosophila allows for the first precise analyses of how global sequence patterns change among multiple, closely related animal species. A basic question is whether there are characteristic features that differentiate chromosomes within a species or between different species.
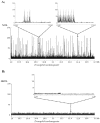
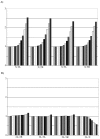
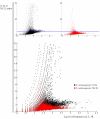
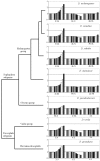
Results: We explored the euchromatin of the chromosomes of seven Drosophila species to establish their global patterns of DNA sequence diversity. Between species, differences in the types and amounts of simple sequence repeats were found. Within each species, the autosomes have almost identical oligonucleotide profiles. However, X chromosomes and autosomes have, in all species, a qualitatively different composition. The X chromosomes are less complex than the autosomes, containing both a higher amount of simple DNA sequences and, in several cases, chromosome-specific repetitive sequences. Moreover, we show that the right arm of the X chromosome of Drosophila pseudoobscura, which evolved from an autosome 10 - 18 millions of years ago, has a composition which is identical to that of the original, left arm of the X chromosome.
Conclusion: The consistent differences among species, differences among X chromosomes and autosomes and the convergent evolution of X and neo-X chromosomes demonstrate that strong forces are acting on drosophilid genomes to generate peculiar chromosomal landscapes. We discuss the relationships of the patterns observed with differential recombination and mutation rates and with the process of dosage compensation.
Figures


Similar articles
-
Gene content evolution on the X chromosome.Curr Opin Genet Dev. 2008 Dec;18(6):493-8. doi: 10.1016/j.gde.2008.09.006. Epub 2008 Oct 16. Curr Opin Genet Dev. 2008. PMID: 18929654 Free PMC article. Review.
-
Adaptive evolution of genes duplicated from the Drosophila pseudoobscura neo-X chromosome.Mol Biol Evol. 2010 Aug;27(8):1963-78. doi: 10.1093/molbev/msq085. Epub 2010 Mar 29. Mol Biol Evol. 2010. PMID: 20351054 Free PMC article.
-
Molecular evolution of the Sex-Ratio inversion complex in Drosophila pseudoobscura: analysis of the Esterase-5 gene region.Mol Biol Evol. 1996 Feb;13(2):297-308. doi: 10.1093/oxfordjournals.molbev.a025589. Mol Biol Evol. 1996. PMID: 8587496
-
Sequence differentiation associated with an inversion on the neo-X chromosome of Drosophila americana.Genetics. 2003 Nov;165(3):1317-28. doi: 10.1093/genetics/165.3.1317. Genetics. 2003. PMID: 14668385 Free PMC article.
-
Evolution of gene function on the X chromosome versus the autosomes.Genome Dyn. 2007;3:101-118. doi: 10.1159/000107606. Genome Dyn. 2007. PMID: 18753787 Review.
Cited by
-
Gene content evolution on the X chromosome.Curr Opin Genet Dev. 2008 Dec;18(6):493-8. doi: 10.1016/j.gde.2008.09.006. Epub 2008 Oct 16. Curr Opin Genet Dev. 2008. PMID: 18929654 Free PMC article. Review.
-
The life cycle of Drosophila orphan genes.Elife. 2014 Feb 19;3:e01311. doi: 10.7554/eLife.01311. Elife. 2014. PMID: 24554240 Free PMC article.
-
Drosophila dosage compensation: males are from Mars, females are from Venus.Fly (Austin). 2011 Apr-Jun;5(2):147-54. doi: 10.4161/fly.5.2.14934. Epub 2011 Apr 1. Fly (Austin). 2011. PMID: 21339706 Free PMC article. Review.
-
A sequence motif enriched in regions bound by the Drosophila dosage compensation complex.BMC Genomics. 2010 Mar 12;11:169. doi: 10.1186/1471-2164-11-169. BMC Genomics. 2010. PMID: 20226017 Free PMC article.
-
Variable Rates of Simple Satellite Gains across the Drosophila Phylogeny.Mol Biol Evol. 2018 Apr 1;35(4):925-941. doi: 10.1093/molbev/msy005. Mol Biol Evol. 2018. PMID: 29361128 Free PMC article.
References
-
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley HC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of Drosophila pseudoobscura : chromosomal, gene, and cis-elements evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

