A Hidden Markov Model to estimate population mixture and allelic copy-numbers in cancers using Affymetrix SNP arrays
- PMID: 17996079
- PMCID: PMC2206057
- DOI: 10.1186/1471-2105-8-434
A Hidden Markov Model to estimate population mixture and allelic copy-numbers in cancers using Affymetrix SNP arrays
Abstract
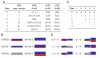
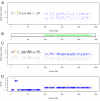
Background: Affymetrix SNP arrays can interrogate thousands of SNPs at the same time. This allows us to look at the genomic content of cancer cells and to investigate the underlying events leading to cancer. Genomic copy-numbers are today routinely derived from SNP array data, but the proposed algorithms for this task most often disregard the genotype information available from germline cells in paired germline-tumour samples. Including this information may deepen our understanding of the "true" biological situation e.g. by enabling analysis of allele specific copy-numbers. Here we rely on matched germline-tumour samples and have developed a Hidden Markov Model (HMM) to estimate allelic copy-number changes in tumour cells. Further with this approach we are able to estimate the proportion of normal cells in the tumour (mixture proportion).
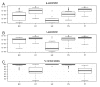
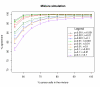
Results: We show that our method is able to recover the underlying copy-number changes in simulated data sets with high accuracy (above 97.71%). Moreover, although the known copy-numbers could be well recovered in simulated cancer samples with more than 70% cancer cells (and less than 30% normal cells), we demonstrate that including the mixture proportion in the HMM increases the accuracy of the method. Finally, the method is tested on HapMap samples and on bladder and prostate cancer samples.
Conclusion: The HMM method developed here uses the genotype calls of germline DNA and the allelic SNP intensities from the tumour DNA to estimate allelic copy-numbers (including changes) in the tumour. It differentiates between different events like uniparental disomy and allelic imbalances. Moreover, the HMM can estimate the mixture proportion, and thus inform about the purity of the tumour sample.
Figures

References
-
- The NCBI dbSNP database http://www.ncbi.nlm.nih.gov/projects/SNP/index.html
-
- Shen R, Fan JB, Campbell D, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Wickham Garcia E, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

