Immobility, inheritance and plasticity of shape of the yeast nucleus
- PMID: 17996101
- PMCID: PMC2222239
- DOI: 10.1186/1471-2121-8-47
Immobility, inheritance and plasticity of shape of the yeast nucleus
Abstract
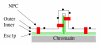
Background: Since S. cerevisiae undergoes closed mitosis, the nuclear envelope of the daughter nucleus is continuous with that of the maternal nucleus at anaphase. Nevertheless, several constitutents of the maternal nucleus are not present in the daughter nucleus. The present study aims to identify proteins which impact the shape of the yeast nucleus and to learn whether modifications of shape are passed on to the next mitotic generation. The Esc1p protein of S. cerevisiae localizes to the periphery of the nucleoplasm, can anchor chromatin, and has been implicated in targeted silencing both at telomeres and at HMR.
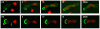
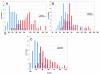
Results: Upon increased Esc1p expression, cell division continues and dramatic elaborations of the nuclear envelope extend into the cytoplasm. These "escapades" include nuclear pores and associate with the nucleolus, but exclude chromatin. Escapades are not inherited by daughter nuclei. This exclusion reflects their relative immobility, which we document in studies of prezygotes. Moreover, excess Esc1p affects the levels of multiple transcripts, not all of which originate at telomere-proximal loci. Unlike Esc1p and the colocalizing protein, Mlp1p, overexpression of selected proteins of the inner nuclear membrane is toxic.
Conclusion: Esc1p is the first non-membrane protein of the nuclear periphery which - like proteins of the nuclear lamina of higher eukaryotes - can modify the shape of the yeast nucleus. The elaborations of the nuclear envelope ("escapades") which appear upon induction of excess Esc1p are not inherited during mitotic growth. The lack of inheritance of such components could help sustain cell growth when parental nuclei have acquired potentially deleterious characteristics.
Figures





References
-
- Loidl J. Chromosomes of the budding yeast Saccharomyces cerevisiae. Int Rev Cytol. 2003;222:141–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

