Structure and evolution of gene regulatory networks in microbial genomes
- PMID: 17996425
- PMCID: PMC5696542
- DOI: 10.1016/j.resmic.2007.09.001
Structure and evolution of gene regulatory networks in microbial genomes
Abstract
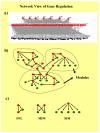
With the availability of genome sequences for hundreds of microbial genomes, it has become possible to address several questions from a comparative perspective to understand the structure and function of regulatory systems, at least in model organisms. Recent studies have focused on topological properties and the evolution of regulatory networks and their components. Our understanding of natural networks is paving the way to embedding synthetic regulatory systems into organisms, allowing us to expand the natural diversity of living systems to an extent we had never before anticipated.
Figures

Similar articles
-
Transcription Factors Exhibit Differential Conservation in Bacteria with Reduced Genomes.PLoS One. 2016 Jan 14;11(1):e0146901. doi: 10.1371/journal.pone.0146901. eCollection 2016. PLoS One. 2016. PMID: 26766575 Free PMC article.
-
Signal correlations in ecological niches can shape the organization and evolution of bacterial gene regulatory networks.Adv Microb Physiol. 2012;61:1-36. doi: 10.1016/B978-0-12-394423-8.00001-9. Adv Microb Physiol. 2012. PMID: 23046950 Free PMC article. Review.
-
In silico simulations of occurrence of transcription factor binding sites in bacterial genomes.BMC Evol Biol. 2019 Mar 1;19(1):67. doi: 10.1186/s12862-019-1381-8. BMC Evol Biol. 2019. PMID: 30823869 Free PMC article.
-
Transcription activation in bacteria: ancient and modern.Microbiology (Reading). 2019 Apr;165(4):386-395. doi: 10.1099/mic.0.000783. Epub 2019 Feb 18. Microbiology (Reading). 2019. PMID: 30775965 Review.
-
RegPrecise 3.0--a resource for genome-scale exploration of transcriptional regulation in bacteria.BMC Genomics. 2013 Nov 1;14:745. doi: 10.1186/1471-2164-14-745. BMC Genomics. 2013. PMID: 24175918 Free PMC article.
Cited by
-
Adaptive Evolution of Thermotoga maritima Reveals Plasticity of the ABC Transporter Network.Appl Environ Microbiol. 2015 Aug 15;81(16):5477-85. doi: 10.1128/AEM.01365-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048924 Free PMC article.
-
Evolution of gene regulatory networks by fluctuating selection and intrinsic constraints.PLoS Comput Biol. 2010 Aug 5;6(8):e1000873. doi: 10.1371/journal.pcbi.1000873. PLoS Comput Biol. 2010. PMID: 20700492 Free PMC article.
-
Identification and genomic analysis of transcription factors in archaeal genomes exemplifies their functional architecture and evolutionary origin.Mol Biol Evol. 2010 Jun;27(6):1449-59. doi: 10.1093/molbev/msq033. Epub 2010 Feb 1. Mol Biol Evol. 2010. PMID: 20123795 Free PMC article.
-
The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii.Nucleic Acids Res. 2012 May;40(10):4306-19. doi: 10.1093/nar/gks014. Epub 2012 Jan 24. Nucleic Acids Res. 2012. PMID: 22275524 Free PMC article.
-
Dissecting the expression patterns of transcription factors across conditions using an integrated network-based approach.Nucleic Acids Res. 2010 Nov;38(20):6841-56. doi: 10.1093/nar/gkq612. Epub 2010 Jul 14. Nucleic Acids Res. 2010. PMID: 20631006 Free PMC article.
References
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
-
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. - PubMed
-
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. - PubMed
-
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

