Structure and evolution of gene regulatory networks in microbial genomes
- PMID: 17996425
- PMCID: PMC5696542
- DOI: 10.1016/j.resmic.2007.09.001
Structure and evolution of gene regulatory networks in microbial genomes
Abstract
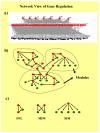
With the availability of genome sequences for hundreds of microbial genomes, it has become possible to address several questions from a comparative perspective to understand the structure and function of regulatory systems, at least in model organisms. Recent studies have focused on topological properties and the evolution of regulatory networks and their components. Our understanding of natural networks is paving the way to embedding synthetic regulatory systems into organisms, allowing us to expand the natural diversity of living systems to an extent we had never before anticipated.
Figures

References
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. - PubMed
-
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. - PubMed
-
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. - PubMed
-
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

