Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design
- PMID: 17996741
- PMCID: PMC2561045
- DOI: 10.1016/j.bbapap.2007.10.002
Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design
Abstract
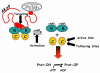
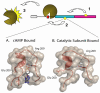
The catalytic subunit of cAMP-dependent protein kinase has served as a prototype for the protein kinase superfamily for many years while structures of the cAMP-bound regulatory subunits have defined the conserved cyclic nucleotide binding (CNB) motif. It is only structures of the holoenzymes, however, that enable us to appreciate the molecular features of inhibition by the regulatory subunits as well as activation by cAMP. These structures reveal for the first time the remarkable malleability of the regulatory subunits and the CNB domains. At the same time, they allow us to appreciate that the catalytic subunit is not only a catalyst but also a scaffold that mediates a wide variety of protein:protein interactions. The holoenzyme structures also provide a new paradigm for designing isoform-specific activators and inhibitors of PKA. In addition to binding to the catalytic subunits, the regulatory subunits also use their N-terminal dimerization/docking domain to bind with high affinity to A Kinase Anchoring Proteins using an amphipathic helical motif. This targeting mechanism, which localizes PKA near to its protein substrates, is also a target for therapeutic intervention of PKA signaling.
Figures

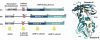





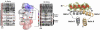
References
-
- Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–5. - PubMed
-
- Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–14. - PubMed
-
- Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9:273–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

