APRIN is a unique Pds5 paralog with features of a chromatin regulator in hormonal differentiation
- PMID: 17997301
- PMCID: PMC3966471
- DOI: 10.1016/j.jsbmb.2007.05.034
APRIN is a unique Pds5 paralog with features of a chromatin regulator in hormonal differentiation
Abstract
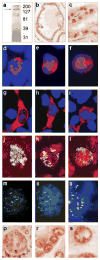
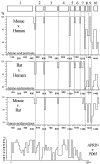
Activation of steroid receptors results in global changes of gene expression patterns. Recent studies showed that steroid receptors control only a portion of their target genes directly, by promoter binding. The majority of the changes are indirect, through chromatin rearrangements. The mediators that relay the hormonal signals to large-scale chromatin changes are, however, unknown. We report here that APRIN, a novel hormone-induced nuclear phosphoprotein has the characteristics of a chromatin regulator and may link endocrine pathways to chromatin. We showed earlier that APRIN is involved in the hormonal regulation of proliferative arrest in cancer cells. To investigate its function we cloned and characterized APRIN orthologs and performed homology and expression studies. APRIN is a paralog of the cohesin-associated Pds5 gene lineage and arose by gene-duplication in early vertebrates. The conservation and domain differences we found suggest, however, that APRIN acquired novel chromatin-related functions (e.g. the HMG-like domains in APRIN, the hallmarks of chromatin regulators, are absent in the Pds5 family). Our results suggest that in interphase nuclei APRIN localizes in the euchromatin/heterochromatin interface and we also identified its DNA-binding and nuclear import signal domains. The results indicate that APRIN, in addition to its Pds5 similarity, has the features and localization of a hormone-induced chromatin regulator.
Figures




Similar articles
-
Loss of a cohesin-linked suppressor APRIN (Pds5b) disrupts stem cell programs in embryonal carcinoma: an emerging cohesin role in tumor suppression.Oncogene. 2010 Jun 10;29(23):3446-52. doi: 10.1038/onc.2010.100. Epub 2010 Apr 12. Oncogene. 2010. PMID: 20383194
-
Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction.Nucleic Acids Res. 2016 Dec 15;44(22):10879-10897. doi: 10.1093/nar/gkw921. Epub 2016 Oct 24. Nucleic Acids Res. 2016. PMID: 27924011 Free PMC article.
-
Arabidopsis chromatin-associated HMGA and HMGB use different nuclear targeting signals and display highly dynamic localization within the nucleus.Plant Cell. 2006 Nov;18(11):2904-18. doi: 10.1105/tpc.106.047274. Epub 2006 Nov 17. Plant Cell. 2006. PMID: 17114349 Free PMC article.
-
Differentiation primary response genes and proto-oncogenes as positive and negative regulators of terminal hematopoietic cell differentiation.Stem Cells. 1994 Jul;12(4):352-69. doi: 10.1002/stem.5530120402. Stem Cells. 1994. PMID: 7951003 Review.
-
The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins.Cell Mol Life Sci. 2007 Oct;64(19-20):2590-606. doi: 10.1007/s00018-007-7162-3. Cell Mol Life Sci. 2007. PMID: 17599239 Free PMC article. Review.
Cited by
-
Nuclear functions of the HMG proteins.Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):3-14. doi: 10.1016/j.bbagrm.2009.09.001. Epub 2009 Sep 11. Biochim Biophys Acta. 2010. PMID: 19748605 Free PMC article. Review.
-
Overexpression of APRIN inhibits differentiation and proliferation and promotes apoptosis in P19 embryonal carcinoma cells.Mol Biol Rep. 2013 Jan;40(1):491-5. doi: 10.1007/s11033-012-2085-y. Epub 2012 Oct 11. Mol Biol Rep. 2013. PMID: 23054015
-
Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses.Endocr Rev. 2012 Jun;33(3):378-455. doi: 10.1210/er.2011-1050. Epub 2012 Mar 14. Endocr Rev. 2012. PMID: 22419778 Free PMC article. Review.
-
Pds5 regulators segregate cohesion and condensation pathways in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):7021-6. doi: 10.1073/pnas.1501369112. Epub 2015 May 18. Proc Natl Acad Sci U S A. 2015. PMID: 25986377 Free PMC article.
-
PDS5B positively regulates PTEN by binding to YTHDC1 to inhibit the malignant progression of endometrial carcinoma.Discov Oncol. 2025 Jul 1;16(1):1192. doi: 10.1007/s12672-025-03021-0. Discov Oncol. 2025. PMID: 40591192 Free PMC article.
References
-
- Sonnenschein C, Soto AM. The Society of Cells: Cancer and Control of Cell Proliferation. Springer Verlag; New York: 1999.
-
- Takahashi K, Ohmichi M, Yoshida M, Hisamoto K, Mabuchi S, Arimoto-Ishida E, Mori A, Tsutsumi S, Tasaka K, Murata Y, Kurachi H. Both estrogen and raloxifene cause G1 arrest of vascular smooth muscle cells. J Endocrinol. 2003;178:319–329. - PubMed
-
- Kofler R, Schmidt S, Kofler A, Ausserlechner MJ. Resistance to glucocorticoid-induced apoptosis in lymphoblastic leukemia. J Endocrinol. 2003;178:19–27. - PubMed
-
- Ling MT, Chan KW, Choo CK. Androgen induces differentiation of a human papillomavirus 16 E6/E7 immortalized prostate epithelial cell line. J Endocrinol. 2001;170:287–296. - PubMed
-
- Udayakumar TS, Jeyaraj DA, Rajalakshmi M, Sharma RS. Culture of prostate epithelial cells of the rhesus monkey on extracellular matrix substrate: influence of steroids and insulin-like growth factors. J Endocrinol. 1999;162:443–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

