A nucleosome-guided map of transcription factor binding sites in yeast
- PMID: 17997593
- PMCID: PMC2065891
- DOI: 10.1371/journal.pcbi.0030215
A nucleosome-guided map of transcription factor binding sites in yeast
Abstract
Finding functional DNA binding sites of transcription factors (TFs) throughout the genome is a crucial step in understanding transcriptional regulation. Unfortunately, these binding sites are typically short and degenerate, posing a significant statistical challenge: many more matches to known TF motifs occur in the genome than are actually functional. However, information about chromatin structure may help to identify the functional sites. In particular, it has been shown that active regulatory regions are usually depleted of nucleosomes, thereby enabling TFs to bind DNA in those regions. Here, we describe a novel motif discovery algorithm that employs an informative prior over DNA sequence positions based on a discriminative view of nucleosome occupancy. When a Gibbs sampling algorithm is applied to yeast sequence-sets identified by ChIP-chip, the correct motif is found in 52% more cases with our informative prior than with the commonly used uniform prior. This is the first demonstration that nucleosome occupancy information can be used to improve motif discovery. The improvement is dramatic, even though we are using only a statistical model to predict nucleosome occupancy; we expect our results to improve further as high-resolution genome-wide experimental nucleosome occupancy data becomes increasingly available.
Conflict of interest statement
Figures


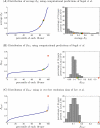
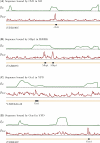


References
-
- Ren B, Robert F, Wyrick J, Aparicio O, Jennings E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

