T cell responses to human endogenous retroviruses in HIV-1 infection
- PMID: 17997601
- PMCID: PMC2065876
- DOI: 10.1371/journal.ppat.0030165
T cell responses to human endogenous retroviruses in HIV-1 infection
Abstract
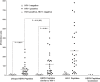
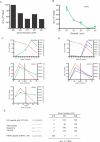
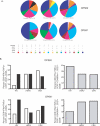
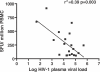
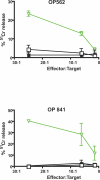
Human endogenous retroviruses (HERVs) are remnants of ancient infectious agents that have integrated into the human genome. Under normal circumstances, HERVs are functionally defective or controlled by host factors. In HIV-1-infected individuals, intracellular defense mechanisms are compromised. We hypothesized that HIV-1 infection would remove or alter controls on HERV activity. Expression of HERV could potentially stimulate a T cell response to HERV antigens, and in regions of HIV-1/HERV similarity, these T cells could be cross-reactive. We determined that the levels of HERV production in HIV-1-positive individuals exceed those of HIV-1-negative controls. To investigate the impact of HERV activity on specific immunity, we examined T cell responses to HERV peptides in 29 HIV-1-positive and 13 HIV-1-negative study participants. We report T cell responses to peptides derived from regions of HERV detected by ELISPOT analysis in the HIV-1-positive study participants. We show an inverse correlation between anti-HERV T cell responses and HIV-1 plasma viral load. In HIV-1-positive individuals, we demonstrate that HERV-specific T cells are capable of killing cells presenting their cognate peptide. These data indicate that HIV-1 infection leads to HERV expression and stimulation of a HERV-specific CD8+ T cell response. HERV-specific CD8+ T cells have characteristics consistent with an important role in the response to HIV-1 infection: a phenotype similar to that of T cells responding to an effectively controlled virus (cytomegalovirus), an inverse correlation with HIV-1 plasma viral load, and the ability to lyse cells presenting their target peptide. These characteristics suggest that elicitation of anti-HERV-specific immune responses is a novel approach to immunotherapeutic vaccination. As endogenous retroviral sequences are fixed in the human genome, they provide a stable target, and HERV-specific T cells could recognize a cell infected by any HIV-1 viral variant. HERV-specific immunity is an important new avenue for investigation in HIV-1 pathogenesis and vaccine design.
Conflict of interest statement
Figures


Similar articles
-
Human endogenous retrovirus expression is inversely associated with chronic immune activation in HIV-1 infection.PLoS One. 2012;7(8):e41021. doi: 10.1371/journal.pone.0041021. Epub 2012 Aug 7. PLoS One. 2012. PMID: 22879884 Free PMC article.
-
Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection.J Virol. 2011 Jul;85(14):6977-85. doi: 10.1128/JVI.00179-11. Epub 2011 Apr 27. J Virol. 2011. PMID: 21525339 Free PMC article.
-
HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates.J Clin Invest. 2012 Dec;122(12):4473-89. doi: 10.1172/JCI64560. Epub 2012 Nov 12. J Clin Invest. 2012. PMID: 23143309 Free PMC article.
-
HIV infection and HERV expression: a review.Retrovirology. 2012 Jan 16;9:6. doi: 10.1186/1742-4690-9-6. Retrovirology. 2012. PMID: 22248111 Free PMC article. Review.
-
Contribution of Human Retroviruses to Disease Development-A Focus on the HIV- and HERV-Cancer Relationships and Treatment Strategies.Viruses. 2020 Aug 4;12(8):852. doi: 10.3390/v12080852. Viruses. 2020. PMID: 32759845 Free PMC article. Review.
Cited by
-
Human endogenous retrovirus expression is inversely associated with chronic immune activation in HIV-1 infection.PLoS One. 2012;7(8):e41021. doi: 10.1371/journal.pone.0041021. Epub 2012 Aug 7. PLoS One. 2012. PMID: 22879884 Free PMC article.
-
Increased seroreactivity to HERV-K10 peptides in patients with HTLV myelopathy.Virol J. 2013 Dec 23;10:360. doi: 10.1186/1743-422X-10-360. Virol J. 2013. PMID: 24365054 Free PMC article.
-
Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis.Ann Neurol. 2011 Jan;69(1):141-51. doi: 10.1002/ana.22149. Ann Neurol. 2011. PMID: 21280084 Free PMC article.
-
Comprehensive Analysis of HERV Transcriptome in HIV+ Cells: Absence of HML2 Activation and General Downregulation of Individual HERV Loci.Viruses. 2020 Apr 23;12(4):481. doi: 10.3390/v12040481. Viruses. 2020. PMID: 32340287 Free PMC article.
-
Novel translation products from simian immunodeficiency virus SIVmac239 Env-encoding mRNA contain both Rev and cryptic T-cell epitopes.J Virol. 2009 Oct;83(19):10280-5. doi: 10.1128/JVI.00138-09. Epub 2009 Jul 15. J Virol. 2009. PMID: 19605480 Free PMC article.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

