Gene family evolution across 12 Drosophila genomes
- PMID: 17997610
- PMCID: PMC2065885
- DOI: 10.1371/journal.pgen.0030197
Gene family evolution across 12 Drosophila genomes
Abstract
Comparison of whole genomes has revealed large and frequent changes in the size of gene families. These changes occur because of high rates of both gene gain (via duplication) and loss (via deletion or pseudogenization), as well as the evolution of entirely new genes. Here we use the genomes of 12 fully sequenced Drosophila species to study the gain and loss of genes at unprecedented resolution. We find large numbers of both gains and losses, with over 40% of all gene families differing in size among the Drosophila. Approximately 17 genes are estimated to be duplicated and fixed in a genome every million years, a rate on par with that previously found in both yeast and mammals. We find many instances of extreme expansions or contractions in the size of gene families, including the expansion of several sex- and spermatogenesis-related families in D. melanogaster that also evolve under positive selection at the nucleotide level. Newly evolved gene families in our dataset are associated with a class of testes-expressed genes known to have evolved de novo in a number of cases. Gene family comparisons also allow us to identify a number of annotated D. melanogaster genes that are unlikely to encode functional proteins, as well as to identify dozens of previously unannotated D. melanogaster genes with conserved homologs in the other Drosophila. Taken together, our results demonstrate that the apparent stasis in total gene number among species has masked rapid turnover in individual gene gain and loss. It is likely that this genomic revolving door has played a large role in shaping the morphological, physiological, and metabolic differences among species.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
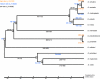
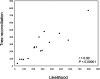
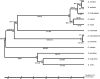
References
-
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. - PubMed
-
- Roelofs J, Van Haastert PJM. Genes lost during evolution. Nature. 2001;411:1013–1014. - PubMed
-
- Hughes AL, Friedman R. Shedding genomic ballast: extensive parallel loss of ancestral gene families in animals. J Mol Evol. 2004;59:827–833. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

