The distributions, mechanisms, and structures of metabolite-binding riboswitches
- PMID: 17997835
- PMCID: PMC2258182
- DOI: 10.1186/gb-2007-8-11-r239
The distributions, mechanisms, and structures of metabolite-binding riboswitches
Abstract
Background: Riboswitches are noncoding RNA structures that appropriately regulate genes in response to changing cellular conditions. The expression of many proteins involved in fundamental metabolic processes is controlled by riboswitches that sense relevant small molecule ligands. Metabolite-binding riboswitches that recognize adenosylcobalamin (AdoCbl), thiamin pyrophosphate (TPP), lysine, glycine, flavin mononucleotide (FMN), guanine, adenine, glucosamine-6-phosphate (GlcN6P), 7-aminoethyl 7-deazaguanine (preQ1), and S-adenosylmethionine (SAM) have been reported.
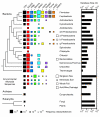
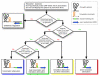
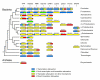
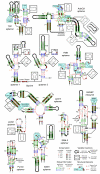
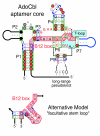
Results: We have used covariance model searches to identify examples of ten widespread riboswitch classes in the genomes of organisms from all three domains of life. This data set rigorously defines the phylogenetic distributions of these riboswitch classes and reveals how their gene control mechanisms vary across different microbial groups. By examining the expanded aptamer sequence alignments resulting from these searches, we have also re-evaluated and refined their consensus secondary structures. Updated riboswitch structure models highlight additional RNA structure motifs, including an unusual double T-loop arrangement common to AdoCbl and FMN riboswitch aptamers, and incorporate new, sometimes noncanonical, base-base interactions predicted by a mutual information analysis.
Conclusion: Riboswitches are vital components of many genomes. The additional riboswitch variants and updated aptamer structure models reported here will improve future efforts to annotate these widespread regulatory RNAs in genomic sequences and inform ongoing structural biology efforts. There remain significant questions about what physiological and evolutionary forces influence the distributions and mechanisms of riboswitches and about what forms of regulation substitute for riboswitches that appear to be missing in certain lineages.
Figures

References
-
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. - PubMed
-
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. - PubMed
-
- Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch Microbiol. 2005;183:151–159. - PubMed
-
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. - PubMed
-
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

