Structural mapping of protein interactions reveals differences in evolutionary pressures correlated to mRNA level and protein abundance
- PMID: 17997970
- PMCID: PMC2600897
- DOI: 10.1016/j.str.2007.09.010
Structural mapping of protein interactions reveals differences in evolutionary pressures correlated to mRNA level and protein abundance
Abstract
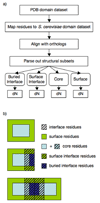
Genome-wide studies in Saccharomyces cerevisiae concluded that the dominant determinant of protein evolutionary rates is expression level: highly expressed proteins generally evolve most slowly. To determine how this constraint affects the evolution of protein interactions, we directly measure evolutionary rates of protein interface, surface, and core residues by structurally mapping domain interactions to yeast genomes. We find that mRNA level and protein abundance, though correlated, report on pressures affecting regions of proteins differently. Pressures proportional to mRNA level slow evolutionary rates of all structural regions and reduce the variability in rate differences between interfaces and other surfaces. In contrast, the evolutionary rate variation within a domain is much less correlated to protein abundance. Distinct pressures may be associated primarily with the cost (mRNA level) and functional (protein abundance) benefit of protein production. Interfaces of proteins with low mRNA levels may have higher evolutionary flexibility and could constitute the raw material for new functions.
Figures





References
-
- Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. Journal of molecular biology. 2003;332:989–998. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

