Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination
- PMID: 17999360
- PMCID: PMC2276354
- DOI: 10.1086/522089
Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination
Abstract
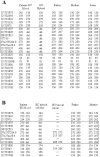
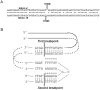
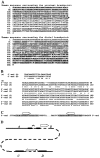
Approximately 5% of patients with neurofibromatosis type 1 (NF1) exhibit gross deletions that encompass the NF1 gene and its flanking regions. The breakpoints of the common 1.4-Mb (type 1) deletions are located within low-copy repeats (NF1-REPs) and cluster within a 3.4-kb hotspot of nonallelic homologous recombination (NAHR). Here, we present the first comprehensive breakpoint analysis of type 2 deletions, which are a second type of recurring NF1 gene deletion. Type 2 deletions span 1.2 Mb and are characterized by breakpoints located within the SUZ12 gene and its pseudogene, which closely flank the NF1-REPs. Breakpoint analysis of 13 independent type 2 deletions did not reveal any obvious hotspots of NAHR. However, an overrepresentation of polypyrimidine/polypurine tracts and triplex-forming sequences was noted in the breakpoint regions that could have facilitated NAHR. Intriguingly, all 13 type 2 deletions identified so far are characterized by somatic mosaicism, which indicates a positional preference for mitotic NAHR within the NF1 gene region. Indeed, whereas interchromosomal meiotic NAHR occurs between the NF1-REPs giving rise to type 1 deletions, NAHR during mitosis appears to occur intrachromosomally between the SUZ12 gene and its pseudogene, thereby generating type 2 deletions. Such a clear distinction between the preferred sites of mitotic versus meiotic NAHR is unprecedented in any other genomic disorder induced by the local genomic architecture. Additionally, 12 of the 13 mosaic type 2 deletions were found in females. The marked female preponderance among mosaic type 2 deletions contrasts with the equal sex distribution noted for type 1 and/or atypical NF1 deletions. Although an influence of chromatin structure was strongly suspected, no sex-specific differences in the methylation pattern exhibited by the SUZ12 gene were apparent that could explain the higher rate of mitotic recombination in females.
Figures
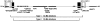

















References
Web Resources
-
- Ensembl, http://www.ensembl.org/index.html (for SUZ12 transcript [accession number ENSG00000178691] and the PanTro 2.1 chimpanzee genome assembly)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SUZ12 [accession number NM_015355], SUZ12P [accession number BC047718], cDNA genomic sequence [accession number AC090616.12], BAC CTD-2349P21 [accession number AC127024], Homo sapiens cDNA clone ESTs [accession numbers BQ185822, AI690568.1, DB451303.1, CX761592.1, BM562953.1, BE166897.1, CN47857.1, and BF817288.1], and BAC RP11-142O6 [accession number AC079915])
-
- HapMap, http://www.hapmap.org/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NF1) - PubMed
-
- Perlegen, http://genome.perlegen.com/
References
-
- Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, de Goede-Bolder A, van den Ouweland AM, Halley DJ, Niermeijer MF (1997) Deletions spanning the neurofibromatosis type 1 gene: implications for genotype-phenotype correlations in neurofibromatosis type 1? Hum Mutat 9:458–46410.1002/(SICI)1098-1004(1997)9:5<458::AID-HUMU13>3.0.CO;2-1 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

