DNA methylation signatures within the human brain
- PMID: 17999367
- PMCID: PMC2276356
- DOI: 10.1086/524110
DNA methylation signatures within the human brain
Abstract
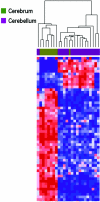
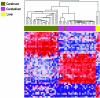
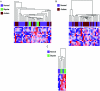
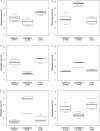
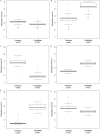
DNA methylation is a heritable modification of genomic DNA central to development, imprinting, transcriptional regulation, chromatin structure, and overall genomic stability. Aberrant DNA methylation of individual genes is a hallmark of cancer and has been shown to play an important role in neurological disorders such as Rett syndrome. Here, we asked whether normal DNA methylation might distinguish individual brain regions. We determined the quantitative DNA methylation levels of 1,505 CpG sites representing 807 genes with diverse functions, including proliferation and differentiation, previously shown to be implicated in human cancer. We initially analyzed 76 brain samples representing cerebral cortex (n=35), cerebellum (n=34), and pons (n=7), along with liver samples (n=3) from 43 individuals. Unsupervised hierarchical analysis showed clustering of 33 of 35 cerebra distinct from the clustering of 33 of 34 cerebella, 7 of 7 pons, and all 3 livers. By use of comparative marker selection and permutation testing, 156 loci representing 118 genes showed statistically significant differences--a >or=17% absolute change in DNA methylation (P<.004)--among brain regions. These results were validated for all six genes tested in a replicate set of 57 samples. Our data suggest that DNA methylation signatures distinguish brain regions and may help account for region-specific functional specialization.
Figures


References
Web Resource
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Rett syndrome, EN2, autism, HDAC7A, HTR2A, SLC22A3, IGF1, FGF1, FGFR2, and MAFD1)
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

