GeneChip analysis of human embryonic stem cell differentiation into hemangioblasts: an in silico dissection of mixed phenotypes
- PMID: 17999768
- PMCID: PMC2258184
- DOI: 10.1186/gb-2007-8-11-r240
GeneChip analysis of human embryonic stem cell differentiation into hemangioblasts: an in silico dissection of mixed phenotypes
Abstract
Background: Microarrays are being used to understand human embryonic stem cell (hESC) differentiation. Most differentiation protocols use a multi-stage approach that induces commitment along a particular lineage. Therefore, each stage represents a more mature and less heterogeneous phenotype. Thus, characterizing the heterogeneous progenitor populations upon differentiation are of increasing importance. Here we describe a novel method of data analysis using a recently developed differentiation protocol involving the formation of functional hemangioblasts from hESCs. Blast cells are multipotent and can differentiate into multiple lineages of hematopoeitic cells (erythroid, granulocyte and macrophage), endothelial and smooth muscle cells.
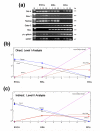
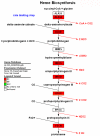
Results: Large-scale transcriptional analysis was performed at distinct time points of hESC differentiation (undifferentiated hESCs, embryoid bodies, and blast cells, the last of which generates both hematopoietic and endothelial progenies). Identifying genes enriched in blast cells relative to hESCs revealed a genetic signature indicative of erythroblasts, suggesting that erythroblasts are the predominant cell type in the blast cell population. Because of the heterogeneity of blast cells, numerous comparisons were made to publicly available data sets in silico, some of which blast cells are capable of differentiating into, to assess and characterize the blast cell population. Biologically relevant comparisons masked particular genetic signatures within the heterogeneous population and identified genetic signatures indicating the presence of endothelia, cardiomyocytes, and hematopoietic lineages in the blast cell population.
Conclusion: The significance of this microarray study is in its ability to assess and identify cellular populations within a heterogeneous population through biologically relevant in silico comparisons of publicly available data sets. In conclusion, multiple in silico comparisons were necessary to characterize tissue-specific genetic signatures within a heterogeneous hemangioblast population.
Figures


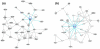
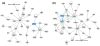
References
-
- Wang L, Li L, Shojaei F, Levac K, Cerdan C, Menendez P, Martin T, Rouleau A, Bhatia Ml. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

