Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks
- PMID: 17999778
- PMCID: PMC2258198
- DOI: 10.1186/gb-2007-8-11-r241
Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks
Abstract
Background: Spore germination in the yeast Saccharomyces cerevisiae is a process in which non-dividing haploid spores re-enter the mitotic cell cycle and resume vegetative growth. To study the signals and pathways underlying spore germination we examined the global changes in gene expression and followed cell-cycle and germination markers during this process.
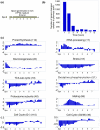
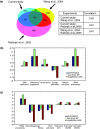
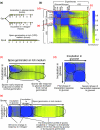
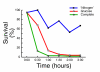
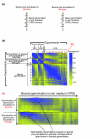
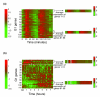
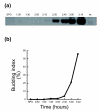
Results: We find that the germination process can be divided into two distinct stages. During the first stage, the induced spores respond only to glucose. The transcription program during this stage recapitulates the general transcription response of yeast cells to glucose. Only during the second phase are the cells able to sense and respond to other nutritional components in the environment. Components of the mitotic machinery are involved in spore germination but in a distinct pattern. In contrast to the mitotic cell cycle, growth-related events during germination are not coordinated with nuclear events and are separately regulated. Thus, genes that are co-induced during G1/S of the mitotic cell cycle, the dynamics of the septin Cdc10 and the kinetics of accumulation of the cyclin Clb2 all exhibit distinct patterns of regulation during spore germination, which allow the separation of cell growth from nuclear events.
Conclusion: Taken together, genome-wide expression profiling enables us to follow the progression of spore germination, thus dividing this process into two major stages, and to identify germination-specific regulation of components of the mitotic cell cycle machinery.
Figures


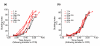
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

