DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI
- PMID: 17999930
- PMCID: PMC2151843
- DOI: 10.1016/j.jmb.2007.10.046
DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI
Abstract
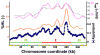
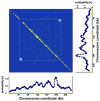
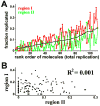
It is generally believed that DNA replication in most eukaryotes proceeds according to a precise program in which there is a defined temporal order by which each chromosomal region is duplicated. However, the regularity of this program at the level of individual chromosomes, in terms of both the relative timing and the size of the DNA domain, has not been addressed. Here, the replication of chromosome VI from synchronized budding yeast was studied at a resolution of approximately 1 kb with DNA combing and fluorescence microscopy. Contrary to what would be expected from cells following a rigorous temporal program, no two molecules exhibited the same replication pattern. Moreover, a direct evaluation of the extent to which the replication of distant chromosomal segments was coordinated indicates that the overwhelming majority of these segments were replicated independently. Importantly, averaging the patterns of all the fibers examined recapitulates the ensemble-averaged patterns obtained from population studies of the replication of chromosome VI. Thus, rather than an absolutely defined temporal order of replication, replication timing appears to be essentially probabilistic within individual cells, exhibiting only temporal tendencies within extended domains.
Figures

References
-
- Goren A, Cedar H. Replicating by the clock. Nat Rev Mol Cell Biol. 2003;4:25–32. - PubMed
-
- Balazs I, Brown EH, Schildkraut CL. The temporal order of replication of some DNA cistrons. Cold Spring Harb Symp Quant Biol. 1974;38:239–45. - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. - PubMed
-
- Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

