Ensembl 2008
- PMID: 18000006
- PMCID: PMC2238821
- DOI: 10.1093/nar/gkm988
Ensembl 2008
Abstract
The Ensembl project (http://www.ensembl.org) is a comprehensive genome information system featuring an integrated set of genome annotation, databases and other information for chordate and selected model organism and disease vector genomes. As of release 47 (October 2007), Ensembl fully supports 35 species, with preliminary support for six additional species. New species in the past year include platypus and horse. Major additions and improvements to Ensembl since our previous report include extensive support for functional genomics data in the form of a specialized functional genomics database, genome-wide maps of protein-DNA interactions and the Ensembl regulatory build; support for customization of the Ensembl web interface through the addition of user accounts and user groups; and increased support for genome resequencing. We have also introduced new comparative genomics-based data mining options and report on the continued development of our software infrastructure.
Figures
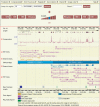

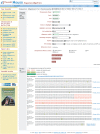

References
-
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
Publication types
MeSH terms
Grants and funding
- BBS/B/13470/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/13462/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E011640/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBE0116401/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 077198/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

