Poxvirus DNA primase
- PMID: 18000036
- PMCID: PMC2141844
- DOI: 10.1073/pnas.0709276104
Poxvirus DNA primase
Abstract
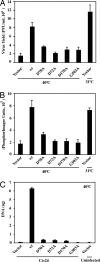
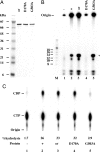
Poxviruses are large enveloped viruses that replicate in the cytoplasm of vertebrate or invertebrate cells. At least six virus-encoded proteins are required for synthesis and processing of the double-stranded DNA genome of vaccinia virus, the prototype member of the family. One of these proteins, D5, is an NTPase that contains an N-terminal archaeoeukaryotic primase domain and a C-terminal superfamily III helicase domain. Here we report that individual conserved aspartic acid residues in the predicted primase active site were required for in vivo complementation of infectious virus formation as well as genome and plasmid replication. Furthermore, purified recombinant D5 protein synthesized oligoribonucleotides in vitro. Incorporation of label from [alpha-(32)P]CTP or [alpha-(32)P]UTP into a RNase-sensitive and DNase-resistant product was demonstrated by using single-stranded circular bacteriophage DNA templates and depended on ATP or GTP and a divalent cation. Mutagenesis studies showed that the primase and NTPase activities of the recombinant D5 protein could be independently inactivated. Highly conserved orthologs of D5 are present in all poxviruses that have been sequenced, and more diverged orthologs are found in members of all other families of nucleocytoplasmic large DNA viruses. These viral primases may have roles in initiation of DNA replication or lagging-strand synthesis and represent potential therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Moss B. In: Fields Virology. Knipe DM, editor. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946.
-
- Moss B, De Silva F. In: DNA Replication and Human Disease. DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 707–727.
-
- Challberg MD, Englund PT. J Biol Chem. 1979;254:7812–7819. - PubMed
-
- McDonald WF, Traktman P. J Biol Chem. 1994;269:31190–31197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

